5WM6
 
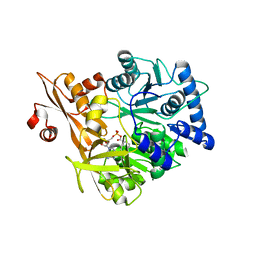 | | Crystal Structure of CahJ in Complex with Benzoyl Adenylate | | Descriptor: | 5'-O-[(R)-(benzoyloxy)(hydroxy)phosphoryl]adenosine, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Sikkema, A.P, Smith, J.L. | | Deposit date: | 2017-07-28 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Defined and Flexible Pocket Explains Aryl Substrate Promiscuity of the Cahuitamycin Starter Unit-Activating Enzyme CahJ.
Chembiochem, 19, 2018
|
|
5WPU
 
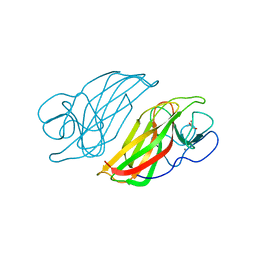 | | Crystal structure HpiC1 Y101S | | Descriptor: | 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
5WPS
 
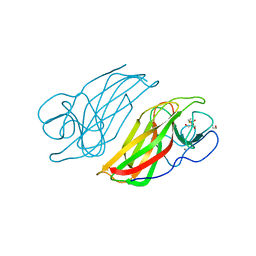 | | Crystal structure HpiC1 Y101F | | Descriptor: | 1,2-ETHANEDIOL, 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.389 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
5WGW
 
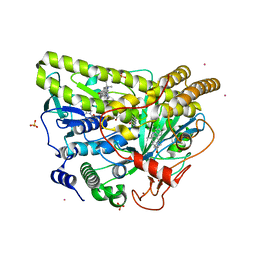 | | Crystal Structure of Wild-type MalA', malbrancheamide B complex | | Descriptor: | (5aS,12aS,13aS)-9-chloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Fraley, A.E, Smith, J.L. | | Deposit date: | 2017-07-14 | | Release date: | 2017-08-16 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Function and Structure of MalA/MalA', Iterative Halogenases for Late-Stage C-H Functionalization of Indole Alkaloids.
J. Am. Chem. Soc., 139, 2017
|
|
5WM7
 
 | | Crystal Structure of CahJ in Complex with AMP | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, GLYCEROL, ... | | Authors: | Sikkema, A.P, Smith, J.L. | | Deposit date: | 2017-07-28 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.777 Å) | | Cite: | A Defined and Flexible Pocket Explains Aryl Substrate Promiscuity of the Cahuitamycin Starter Unit-Activating Enzyme CahJ.
Chembiochem, 19, 2018
|
|
4TPL
 
 | | West Nile Virus Non-structural protein 1 (NS1) Form 1 crystal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, OXTOXYNOL-10, ... | | Authors: | Akey, D.L, Smith, J.L. | | Deposit date: | 2014-06-08 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Use of massively multiple merged data for low-resolution S-SAD phasing and refinement of flavivirus NS1.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6AL7
 
 | | Crystal structure HpiC1 F138S | | Descriptor: | 12-epi-hapalindole C/U synthase, CALCIUM ION | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.687 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
6AL6
 
 | | Crystal structure HpiC1 in P42 space group | | Descriptor: | 12-epi-hapalindole C/U synthase, CALCIUM ION | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.088 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
6AL8
 
 | | Crystal structure HpiC1 Y101F/F138S | | Descriptor: | 1,2-ETHANEDIOL, 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
6BEV
 
 | | Human Single Domain Sulfurtranferase TSTD1 | | Descriptor: | Thiosulfate sulfurtransferase/rhodanese-like domain-containing protein 1 | | Authors: | Motl, N, Akey, D.L, Smith, J.L, Banerjee, R. | | Deposit date: | 2017-10-25 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.043 Å) | | Cite: | Thiosulfate sulfurtransferase-like domain-containing 1 protein interacts with thioredoxin.
J. Biol. Chem., 293, 2018
|
|
3V7I
 
 | |
3SSN
 
 | | MycE Methyltransferase from the Mycinamycin Biosynthetic Pathway in Complex with Mg, SAH, and Mycinamycin VI | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Akey, D.L, Smith, J.L. | | Deposit date: | 2011-07-08 | | Release date: | 2011-08-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | A new structural form in the SAM/metal-dependent o‑methyltransferase family: MycE from the mycinamicin biosynthetic pathway.
J.Mol.Biol., 413, 2011
|
|
3SSO
 
 | |
3SSM
 
 | |
3U1T
 
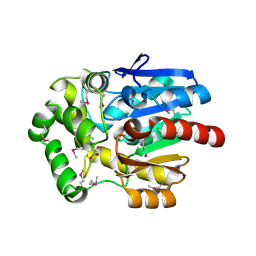 | | Haloalkane Dehalogenase, DmmA, of marine microbial origin | | Descriptor: | CHLORIDE ION, DmmA Haloalkane Dehalogenase, MALONATE ION | | Authors: | Gehret, J.J, Smith, J.L. | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-28 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and activity of DmmA, a marine haloalkane dehalogenase.
Protein Sci., 21, 2012
|
|
9CGL
 
 | | Pikromycin Thioesterase Doubly Protected DAP | | Descriptor: | 2-{[(1R)-1-(6-nitro-2H-1,3-benzodioxol-5-yl)ethyl]sulfanyl}ethyl formate, Narbonolide/10-deoxymethynolide synthase PikA4, module 6 | | Authors: | McCullough, T.M, Smith, J.L. | | Deposit date: | 2024-06-29 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Substrate Trapping in Polyketide Synthase Thioesterase Domains: Structural Basis for Macrolactone Formation
Acs Catalysis, 14, 2024
|
|
9CBD
 
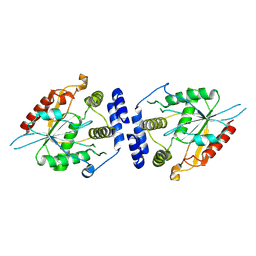 | | Pikromycin Thioesterase Domain | | Descriptor: | Narbonolide/10-deoxymethynolide synthase PikA4, module 6 | | Authors: | McCullough, T.M, Smith, J.L. | | Deposit date: | 2024-06-19 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate Trapping in Polyketide Synthase Thioesterase Domains: Structural Basis for Macrolactone Formation
Acs Catalysis, 14, 2024
|
|
9CFJ
 
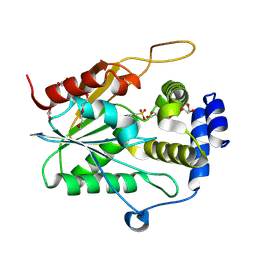 | | Fluvirucin Thioesterase Domain (FluC TE) | | Descriptor: | FluC, GLYCEROL, PENTAETHYLENE GLYCOL | | Authors: | Choudhary, V, Smith, J.L. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Substrate Trapping in Polyketide Synthase Thioesterase Domains: Structural Basis for Macrolactone Formation
Acs Catalysis, 14, 2024
|
|
5K6K
 
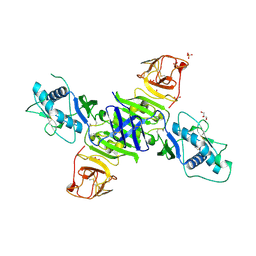 | | Zika virus non-structural protein 1 (NS1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Akey, D.L, Brown, W.C, Smith, J.L. | | Deposit date: | 2016-05-24 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Extended surface for membrane association in Zika virus NS1 structure.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5KP8
 
 | |
5KP5
 
 | |
5KP7
 
 | |
5KP6
 
 | |
4V9E
 
 | |
4X7U
 
 | |
