1K9O
 
 | | CRYSTAL STRUCTURE OF MICHAELIS SERPIN-TRYPSIN COMPLEX | | Descriptor: | ALASERPIN, TRYPSIN II ANIONIC | | Authors: | Ye, S, Cech, A.L, Belmares, R, Bergstrom, R.C, Tong, Y, Corey, D.R, Kanost, M.R, Goldsmith, E.J. | | Deposit date: | 2001-10-29 | | Release date: | 2001-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of a Michaelis serpin-protease complex.
Nat.Struct.Biol., 8, 2001
|
|
1LEW
 
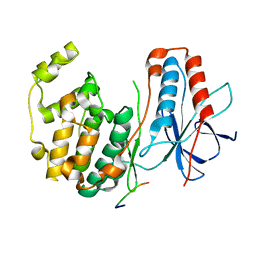 | | CRYSTAL STRUCTURE OF MAP KINASE P38 COMPLEXED TO THE DOCKING SITE ON ITS NUCLEAR SUBSTRATE MEF2A | | Descriptor: | Mitogen-activated protein kinase 14, Myocyte-specific enhancer factor 2A | | Authors: | Chang, C.-I, Xu, B.-E, Akella, R, Cobb, M.H, Goldsmith, E.J. | | Deposit date: | 2002-04-10 | | Release date: | 2002-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b.
Mol.Cell, 9, 2002
|
|
2NV9
 
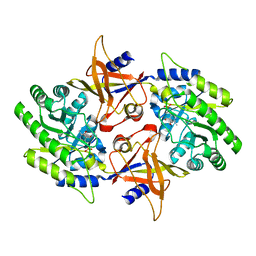 | | The X-ray Crystal Structure of the Paramecium bursaria Chlorella virus arginine decarboxylase | | Descriptor: | A207R protein, arginine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Shah, R.H, Akella, R, Goldsmith, E, Phillips, M.A. | | Deposit date: | 2006-11-11 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray Structure of Paramecium bursaria Chlorella Virus Arginine Decarboxylase: Insight into the Structural Basis for Substrate Specificity.
Biochemistry, 46, 2007
|
|
1NJJ
 
 | |
2NVA
 
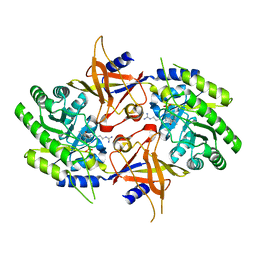 | | The X-ray crystal structure of the Paramecium bursaria Chlorella virus arginine decarboxylase bound to agmatine | | Descriptor: | (4-{[(4-{[AMINO(IMINO)METHYL]AMINO}BUTYL)AMINO]METHYL}-5-HYDROXY-6-METHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, arginine decarboxylase, A207R protein | | Authors: | Shah, R.H, Akella, R, Goldsmith, E, Phillips, M.A. | | Deposit date: | 2006-11-11 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray Structure of Paramecium bursaria Chlorella Virus Arginine Decarboxylase: Insight into the Structural Basis for Substrate Specificity.
Biochemistry, 46, 2007
|
|
1QU4
 
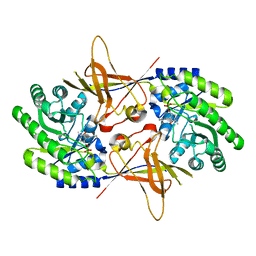 | | CRYSTAL STRUCTURE OF TRYPANOSOMA BRUCEI ORNITHINE DECARBOXYLASE | | Descriptor: | ORNITHINE DECARBOXYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Grishin, N.V, Osterman, A.L, Brooks, H.B, Phillips, M.A, Goldsmith, E.J. | | Deposit date: | 1999-07-06 | | Release date: | 1999-11-17 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray structure of ornithine decarboxylase from Trypanosoma brucei: the native structure and the structure in complex with alpha-difluoromethylornithine.
Biochemistry, 38, 1999
|
|
5UOJ
 
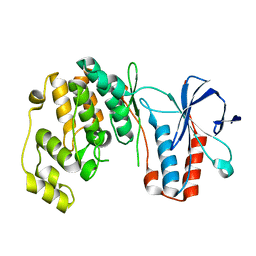 | |
1LEZ
 
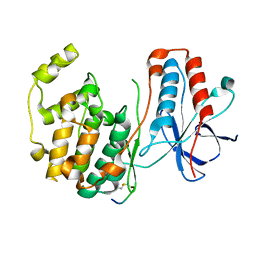 | | CRYSTAL STRUCTURE OF MAP KINASE P38 COMPLEXED TO THE DOCKING SITE ON ITS ACTIVATOR MKK3B | | Descriptor: | MAP kinase kinase 3b, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Chang, C.-I, Xu, B.-E, Akella, R, Cobb, M.H, Goldsmith, E.J. | | Deposit date: | 2002-04-10 | | Release date: | 2002-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b.
Mol.Cell, 9, 2002
|
|
5UMO
 
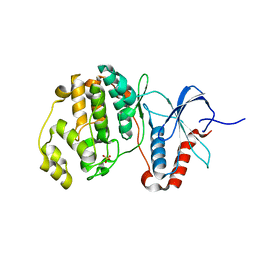 | |
2PLK
 
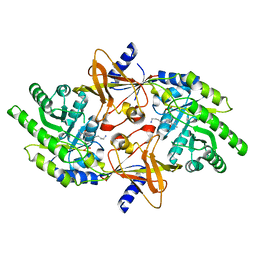 | |
3DAK
 
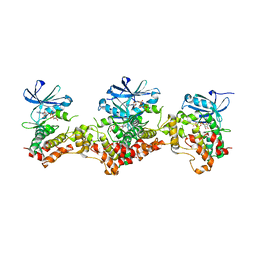 | |
1F3T
 
 | | CRYSTAL STRUCTURE OF TRYPANOSOMA BRUCEI ORNITHINE DECARBOXYLASE (ODC) COMPLEXED WITH PUTRESCINE, ODC'S REACTION PRODUCT. | | Descriptor: | 1,4-DIAMINOBUTANE, ORNITHINE DECARBOXYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jackson, L.K, Brooks, H.B, Osterman, A.L, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2000-06-06 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Altering the reaction specificity of eukaryotic ornithine decarboxylase.
Biochemistry, 39, 2000
|
|
1GOL
 
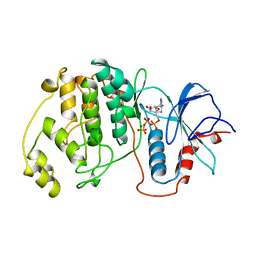 | |
3ENM
 
 | | The structure of the MAP2K MEK6 reveals an autoinhibitory dimer | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity mitogen-activated protein kinase kinase 6, GLYCEROL, ... | | Authors: | Min, X, Akella, R, He, H, Humphreys, J.M, Tsutakawa, S, Lee, S.-J, Tainer, J.A, Cobb, M.H, Goldsmith, E.J. | | Deposit date: | 2008-09-25 | | Release date: | 2009-03-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of the MAP2K MEK6 reveals an autoinhibitory dimer
Structure, 17, 2009
|
|
2PLJ
 
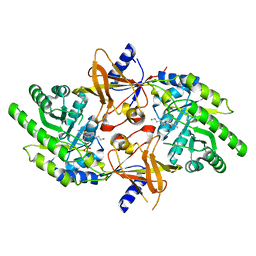 | |
2GCD
 
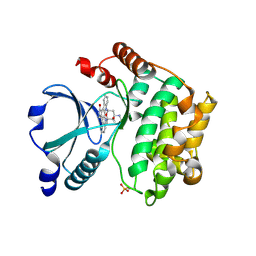 | | TAO2 kinase domain-staurosporine structure | | Descriptor: | STAUROSPORINE, Serine/threonine-protein kinase TAO2 | | Authors: | Zhou, T, Sun, L, Gao, Y, Earnest, S, Cobb, M.H, Goldsmith, E.J. | | Deposit date: | 2006-03-14 | | Release date: | 2006-09-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the MAP3K TAO2 kinase domain bound by an inhibitor staurosporine.
Acta Biochim.Biophys.Sinica, 38, 2006
|
|
2ERK
 
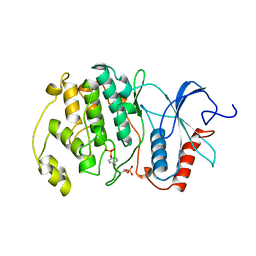 | | PHOSPHORYLATED MAP KINASE ERK2 | | Descriptor: | EXTRACELLULAR SIGNAL-REGULATED KINASE 2 | | Authors: | Canagarajah, B.J, Goldsmith, E.J. | | Deposit date: | 1997-06-26 | | Release date: | 1998-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Activation mechanism of the MAP kinase ERK2 by dual phosphorylation.
Cell(Cambridge,Mass.), 90, 1997
|
|
3ERK
 
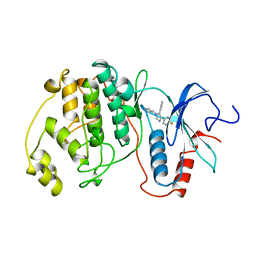 | | THE COMPLEX STRUCTURE OF THE MAP KINASE ERK2/SB220025 | | Descriptor: | 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, EXTRACELLULAR REGULATED KINASE 2 | | Authors: | Wang, Z, Canagarajah, B, Boehm, J.C, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-09 | | Release date: | 1999-07-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
5D9H
 
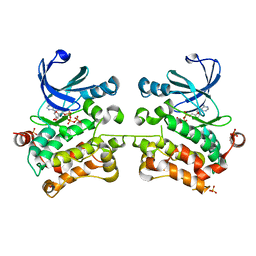 | | Crystal structure of SPAK (STK39) dimer in the basal activity state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, STE20/SPS1-related proline-alanine-rich protein kinase, ... | | Authors: | Taylor, C.A, Juang, Y.C, Goldsmith, E.J, Cobb, M.H. | | Deposit date: | 2015-08-18 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Domain-Swapping Switch Point in Ste20 Protein Kinase SPAK.
Biochemistry, 54, 2015
|
|
3N29
 
 | | Crystal structure of carboxynorspermidine decarboxylase complexed with Norspermidine from Campylobacter jejuni | | Descriptor: | Carboxynorspermidine decarboxylase, GLYCEROL, N-(3-aminopropyl)propane-1,3-diamine, ... | | Authors: | Deng, X, Lee, J, Michael, A.J, Tomchick, D.R, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-09 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of substrate specificity within a diverse family of beta/alpha-barrel-fold basic amino acid decarboxylases: X-ray structure determination of enzymes with specificity for L-arginine and carboxynorspermidine.
J.Biol.Chem., 285, 2010
|
|
3N2O
 
 | | X-ray crystal structure of arginine decarboxylase complexed with Arginine from Vibrio vulnificus | | Descriptor: | AGMATINE, Biosynthetic arginine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Deng, X, Lee, J, Michael, A.J, Tomchick, D.R, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2010-05-18 | | Release date: | 2010-06-09 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of substrate specificity within a diverse family of beta/alpha-barrel-fold basic amino acid decarboxylases: X-ray structure determination of enzymes with specificity for L-arginine and carboxynorspermidine.
J.Biol.Chem., 285, 2010
|
|
3P4K
 
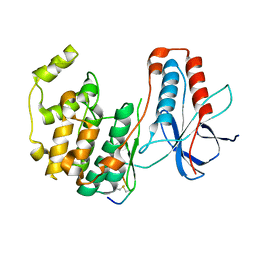 | | The third conformation of p38a MAP kinase observed in phosphorylated p38a and in solution | | Descriptor: | MAP kinase 14, Mitogen-activated protein kinase 14 | | Authors: | Akella, R, Min, X, Wu, Q, Gardner, K.H, Goldsmith, E.J. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | The third conformation of p38a MAP kinase observed in phosphorylated p38a and in solution
Structure, 18, 2010
|
|
2GPH
 
 | | Docking motif interactions in the MAP kinase ERK2 | | Descriptor: | Mitogen-activated protein kinase 1, Tyrosine-protein phosphatase non-receptor type 7 | | Authors: | Zhou, T, Sun, L, Humphreys, J, Goldsmith, E.J. | | Deposit date: | 2006-04-17 | | Release date: | 2006-07-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Docking Interactions Induce Exposure of Activation Loop in the MAP Kinase ERK2.
Structure, 14, 2006
|
|
4ERK
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE ERK2/OLOMOUCINE | | Descriptor: | EXTRACELLULAR REGULATED KINASE 2, OLOMOUCINE, SULFATE ION | | Authors: | Wang, Z, Canagarajah, B, Boehm, J.C, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-09 | | Release date: | 1999-07-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
2LRU
 
 | | Solution Structure of the WNK1 Autoinhibitory Domain | | Descriptor: | Serine/threonine-protein kinase WNK1 | | Authors: | Moon, T.M, Correa, F, Gardner, K.H, Goldsmith, E.J. | | Deposit date: | 2012-04-13 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the WNK1 Autoinhibitory Domain, a WNK-Specific PF2 Domain.
J.Mol.Biol., 425, 2013
|
|
