5T06
 
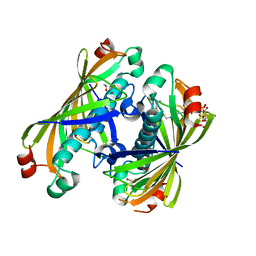 | | Crystal structure of a putative acyl-CoA thioesterase EC709/ECK0725 from Escherichia coli in complex with Hexanoyl-CoA | | Descriptor: | 1,2-ETHANEDIOL, Acyl-CoA thioester hydrolase YbgC, HEXANOYL-COENZYME A | | Authors: | Watanabe, N, Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-15 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal structure of a putative acyl-CoA thioesterase EC709/ECK0725 from Escherichia coli in complex with Hexanoyl-CoA
To be published
|
|
5T07
 
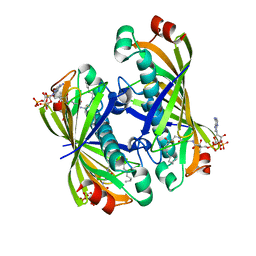 | | Crystal structure of a putative acyl-CoA thioesterase EC709/ECK0725 from Escherichia coli in complex with Decanoyl-CoA | | Descriptor: | Acyl-CoA thioester hydrolase YbgC, decanoyl-CoA | | Authors: | Watanabe, N, Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-15 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | Crystal structure of a putative acyl-CoA thioesterase EC709/ECK0725 from Escherichia coli in complex with Decanoyl-CoA
To be published
|
|
6W4H
 
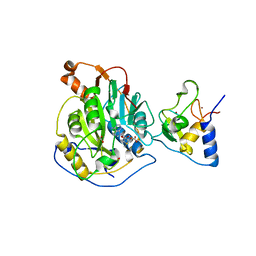 | | 1.80 Angstrom Resolution Crystal Structure of NSP16 - NSP10 Complex from SARS-CoV-2 | | Descriptor: | 2'-O-methyltransferase, ACETATE ION, Non-structural protein 10, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Wiersum, G, Godzik, A, Jaroszewski, L, Stogios, P.J, Skarina, T, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-10 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
4JID
 
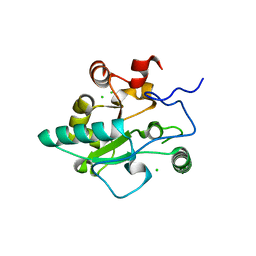 | | Crystal structure of BaLdcB / VanY-like L,D-carboxypeptidase Zinc(II)-free | | Descriptor: | CHLORIDE ION, D-alanyl-D-alanine carboxypeptidase family protein | | Authors: | Minasov, G, Wawrzak, Z, Onopriyenko, O, Skarina, T, Shatsman, S, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-05 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the LdcB LD-Carboxypeptidase Reveals the Molecular Basis of Peptidoglycan Recognition.
Structure, 22, 2014
|
|
6WON
 
 | | Crystal structure of acetoin dehydrogenase YohF from Salmonella typhimurium | | Descriptor: | CHLORIDE ION, SULFATE ION, YohF | | Authors: | Stogios, P.J, Skarina, T, Mesa, N, Endres, M, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-25 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of acetoin dehydrogenase YohF from Salmonella typhimurium
To Be Published
|
|
6WOP
 
 | | Crystal structure of gamma-aminobutyrate aminotransferase PuuE from Acinetobacter baumannii | | Descriptor: | 4-aminobutyrate transaminase, CHLORIDE ION, D(-)-TARTARIC ACID | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-25 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of gamma-aminobutyrate aminotransferase PuuE from Acinetobacter baumannii
To Be Published
|
|
6C8Q
 
 | | Crystal structure of NAD synthetase (NadE) from Enterococcus faecalis in complex with NAD+ | | Descriptor: | NH(3)-dependent NAD(+) synthetase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Stogios, P.J, Skarina, T, McChesney, C, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-01-25 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.583 Å) | | Cite: | To be published
To Be Published
|
|
8EWO
 
 | | Crystal structure of putative glyoxylase II from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, GLYCEROL, PA1813, ... | | Authors: | Stogios, P.J, Skarina, T, Endres, M, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-10-24 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of putative glyoxylase II from Pseudomonas aeruginosa
To Be Published
|
|
7KZW
 
 | | Crystal structure of FTT_1639c from Francisella tularensis str. tularensis SCHU S4 | | Descriptor: | CHLORIDE ION, FTT_1639c | | Authors: | Stogios, P.J, Skarina, T, Osipiuk, J, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-10 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal structure of FTT_1639c from Francisella tularensis str. tularensis SCHU S4
To Be Published
|
|
8EZR
 
 | | Crystal structure of the HipS(Lp)-HipT(Lp) complex from Legionella pneumophila, native protein | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, HipS(Lp), ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Di Leo, R, Lin, J, Ensminger, A, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-11-01 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the HipS(Lp)-HipT(Lp) complex from Legionella pneumophila, native protein
To Be Published
|
|
8EZT
 
 | | Crystal structure of HipB(Lp) from Legionella pneumophila | | Descriptor: | CHLORIDE ION, HipB(Lp) | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Lin, J, Ensminger, A, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-11-01 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of HipB(Lp) from Legionella pneumophila
To Be Published
|
|
8EZS
 
 | | Crystal structure of the HipS(Lp)-HipT(Lp) complex from Legionella pneumophila, Sel-met protein | | Descriptor: | CHLORIDE ION, HipS(Lp), HipT(Lp) | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Di Leo, R, Lin, J, Ensminger, A, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-11-01 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of the HipS(Lp)-HipT(Lp) complex from Legionella pneumophila, Sel-met protein
To Be Published
|
|
8EJV
 
 | | The crystal structure of Pseudomonas putida PcaR in complex with succinate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Pham, C, Skarina, T, Di Leo, R, Stogios, P.J, Mahadevan, R, Savchenko, A. | | Deposit date: | 2022-09-19 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The crystal structure of Pseudomonas putida PcaR in complex with succinate
To Be Published
|
|
7KAG
 
 | | Crystal structure of the ubiquitin-like domain 1 (Ubl1) of Nsp3 from SARS-CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 3, SULFATE ION | | Authors: | Stogios, P.J, Skarina, T, Chang, C, Kim, Y, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-30 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Crystal structure of the ubiquitin-like domain 1 (Ubl1) of Nsp3 from SARS-CoV-2
To Be Published
|
|
7LGO
 
 | | Crystal structure of the nucleic acid binding domain (NAB) of Nsp3 from SARS-CoV-2 | | Descriptor: | Non-structural protein 3 | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-01-20 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the nucleic acid binding domain (NAB) of Nsp3 from SARS-CoV-2
To Be Published
|
|
5HMN
 
 | | Crystal structure of an aminoglycoside acetyltransferase HMB0005 from an uncultured soil metagenomic sample, unknown active site density modeled as polyethylene glycol | | Descriptor: | AAC3-I, COENZYME A, TETRAETHYLENE GLYCOL | | Authors: | Xu, Z, Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-16 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.018 Å) | | Cite: | Crystal structure of an aminoglycoside acetyltransferase HMB0005 from an uncultured soil metagenomic sample, unknown active site density modeled as polyethylene glycol
To Be Published
|
|
7T9W
 
 | | Crystal structure of the Nsp3 bSM (Betacoronavirus-Specific Marker) domain from SARS-CoV-2 | | Descriptor: | CHLORIDE ION, GLYCEROL, Papain-like protease nsp3 | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-20 | | Release date: | 2021-12-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Nsp3 bSM (Betacoronavirus-Specific Marker) domain from SARS-CoV-2
To Be Published
|
|
5IR0
 
 | | Crystal structure of protein of unknown function ORF19 from Vibrio cholerae O1 PICI-like element, C57S I109M mutant | | Descriptor: | CITRIC ACID, Uncharacterized protein ORF19 | | Authors: | Stogios, P.J, Wawrzak, Z, Skarina, T, Di Leo, R, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-03-11 | | Release date: | 2016-03-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.297 Å) | | Cite: | Crystal structure of protein of unknown function ORF19 from Vibrio cholerae O1 PICI-like element, C57S I109M mutant
To Be Published
|
|
6MN1
 
 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in abortive complex with gentamicin-CoA | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Aminoglycoside N(3)-acetyltransferase, ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Xu, Z, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in abortive complex with gentamicin-CoA
To Be Published
|
|
5DT9
 
 | | Crystal structure of a putative D-Erythronate-4-Phosphate Dehydrogenase from Vibrio cholerae | | Descriptor: | CHLORIDE ION, Erythronate-4-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Stogios, P.J, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-17 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.663 Å) | | Cite: | Crystal structure of a putative D-Erythronate-4-Phosphate Dehydrogenase from Vibrio cholerae
To Be Published
|
|
5F13
 
 | | Structure of Mn bound DUF89 from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Nocek, B, Skarina, T, Joachimiak, A, Savchenko, A, Yakunin, A. | | Deposit date: | 2015-11-30 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | A family of metal-dependent phosphatases implicated in metabolite damage-control.
Nat.Chem.Biol., 12, 2016
|
|
5U1H
 
 | | Crystal structure of the C-terminal peptidoglycan binding domain of OprF (PA1777) from Pseudomonas aeruginosa | | Descriptor: | (2R,6S)-2-amino-6-(carboxyamino)-7-{[(1R)-1-carboxyethyl]amino}-7-oxoheptanoic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Watanabe, N, Stogios, P.J, Skarina, T, Wawrzak, Z, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-28 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the C-terminal peptidoglycan binding domain of OprF (PA1777) from Pseudomonas aeruginosa
To be published
|
|
6D2X
 
 | | Crystal structure of the GH26 domain from PbGH26-GH5A endo-beta-mannanase/endo-beta-glucanase from Prevotella bryantii | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Aryl-phospho-beta-D-glucosidase BglC, GH1 family, ... | | Authors: | Stogios, P.J, Skarina, T, McGregor, N, Di Leo, R, Brumer, H, Savchenko, A. | | Deposit date: | 2018-04-14 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of the GH26 domain from PbGH26-GH5A endo-beta-mannanase/endo-beta-glucanase from Prevotella bryantii
To Be Published
|
|
6D2W
 
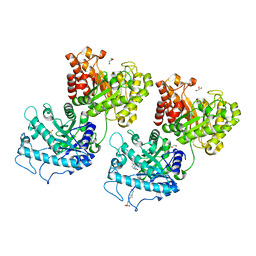 | | Crystal structure of Prevotella bryantii endo-beta-mannanase/endo-beta-glucanase PbGH26A-GH5A | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Aryl-phospho-beta-D-glucosidase BglC, GH1 family, ... | | Authors: | Stogios, P.J, Skarina, T, McGregor, N, Nocek, B, Di Leo, R, Brumer, H, Savchenko, A. | | Deposit date: | 2018-04-14 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Prevotella bryantii endo-beta-mannanase/endo-beta-glucanase PbGH26A-GH5A
To Be Published
|
|
6O62
 
 | | Crystal structure of Sec4p, a Rab family GTPase from Candida albicans | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Ras-related protein SEC4 | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-03-05 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | To be published
To Be Published
|
|
