3LGS
 
 | |
3BL6
 
 | | Crystal structure of Staphylococcus aureus 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase in complex with formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, 5'-methylthioadenosine nucleosidase/S-adenosylhomocysteine nucleosidase | | Authors: | Siu, K.K.W, Lee, J.E, Smith, G.D, Horvatin, C, Howell, P.L. | | Deposit date: | 2007-12-10 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Staphylococcus aureus 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase
Acta Crystallogr.,Sect.F, 64, 2008
|
|
3DF9
 
 | |
2QSU
 
 | |
2QTG
 
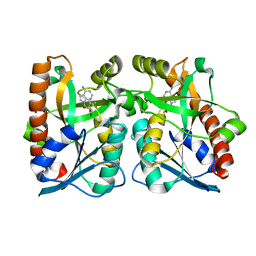 | | Crystal Structure of Arabidopsis thaliana 5'-Methylthioadenosine nucleosidase in complex with 5'-methylthiotubercidin | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-METHYLSULFANYLMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, 5'-methylthioadenosine nucleosidase | | Authors: | Siu, K.K.W, Howell, P.L. | | Deposit date: | 2007-08-02 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Molecular determinants of substrate specificity in plant 5'-methylthioadenosine nucleosidases.
J.Mol.Biol., 378, 2008
|
|
2QTT
 
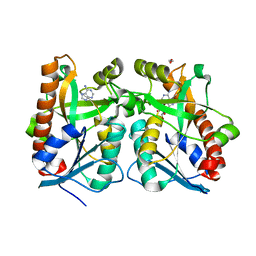 | | Crystal Structure of Arabidopsis thaliana 5'-Methylthioadenosine nucleosidase in complex with Formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, 1,2-ETHANEDIOL, 5'-methylthioadenosine nucleosidase, ... | | Authors: | Siu, K.K.W, Howell, P.L. | | Deposit date: | 2007-08-02 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Molecular determinants of substrate specificity in plant 5'-methylthioadenosine nucleosidases.
J.Mol.Biol., 378, 2008
|
|
3MMS
 
 | | Crystal structure of Streptococcus pneumoniae MTA/SAH nucleosidase in complex with 8-aminoadenine | | Descriptor: | 5'-methylthioadenosine / S-adenosylhomocysteine nucleosidase, 9H-purine-6,8-diamine, GLYCEROL | | Authors: | Siu, K.K.W, Lee, J.E, Horvatin-Mrakovcic, C, Howell, P.L. | | Deposit date: | 2010-04-20 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Streptococcus pneumoniae MTA/SAH nucleosidase in complex with 8-aminoadenine
TO BE PUBLISHED
|
|
3O4V
 
 | |
4J4J
 
 | |
4ID7
 
 | | ACK1 kinase in complex with the inhibitor cis-3-[8-amino-1-(4-phenoxyphenyl)imidazo[1,5-a]pyrazin-3-yl]cyclobutanol | | Descriptor: | Activated CDC42 kinase 1, SULFATE ION, cis-3-[8-amino-1-(4-phenoxyphenyl)imidazo[1,5-a]pyrazin-3-yl]cyclobutanol | | Authors: | Jin, M, Wang, J, Kleinberg, A, Kadalbajoo, M, Siu, K, Cooke, A, Bittner, M, Yao, Y, Thelemann, A, Ji, Q, Bhagwat, S, Crew, A.P, Pachter, J, Epstein, D, Mulvihill, M.J. | | Deposit date: | 2012-12-11 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of potent, selective and orally bioavailable imidazo[1,5-a]pyrazine derived ACK1 inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2V5Q
 
 | | CRYSTAL STRUCTURE OF WILD-TYPE PLK-1 KINASE DOMAIN IN COMPLEX WITH A SELECTIVE DARPIN | | Descriptor: | DESIGN ANKYRIN REPEAT PROTEIN, SERINE/THREONINE-PROTEIN KINASE PLK1 | | Authors: | Bandeiras, T.M, Hillig, R.C, Matias, P.M, Eberspaecher, U, Fanghaenel, J, Thomaz, M, Miranda, S, Crusius, K, Puetter, V, Amstutz, P, Gulotti-Georgieva, M, Binz, H.K, Holz, C, Schmitz, A.A.P, Lang, C, Donner, P, Egner, U, Carrondo, M.A, Mueller-Tiemann, B. | | Deposit date: | 2007-07-08 | | Release date: | 2008-04-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of wild-type Plk-1 kinase domain in complex with a selective DARPin.
Acta Crystallogr. D Biol. Crystallogr., 64, 2008
|
|
1GS4
 
 | | Structural basis for the glucocorticoid response in a mutant human androgen receptor (ARccr) derived from an androgen-independent prostate cancer | | Descriptor: | 9ALPHA-FLUOROCORTISOL, ANDROGEN RECEPTOR, PHOSPHATE ION | | Authors: | Matias, P.M, Carrondo, M.A, Coelho, R, Thomaz, M, Zhao, X.-Y, Wegg, A, Crusius, K, Egner, U, Donner, P. | | Deposit date: | 2001-12-27 | | Release date: | 2003-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for the Glucocorticoid Response in a Mutant Human Androgen Receptor (Ar(Ccr)) Derived from an Androgen-Independent Prostate Cancer
J.Med.Chem., 45, 2002
|
|
