1G0Z
 
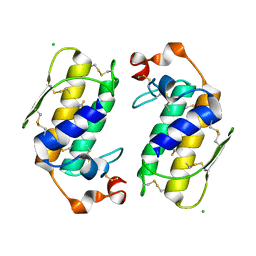 | |
1BJR
 
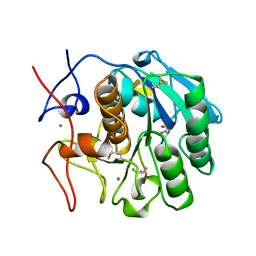 | | COMPLEX FORMED BETWEEN PROTEOLYTICALLY GENERATED LACTOFERRIN FRAGMENT AND PROTEINASE K | | Descriptor: | CALCIUM ION, LACTOFERRIN, PROTEINASE K | | Authors: | Singh, T.P, Sharma, S, Karthikeyan, S, Betzel, C, Bhatia, K.L. | | Deposit date: | 1998-06-27 | | Release date: | 1998-11-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of a complex formed between proteolytically-generated lactoferrin fragment and proteinase K.
Proteins, 33, 1998
|
|
3K0V
 
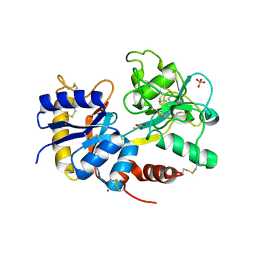 | | Removal of sugars and sugars-like molecules from the solution by C-lobe of lactoferrin: Crystal structure of the complex of C-lobe with beta-D-glucopyranosyl-(1->4)-beta-D-galactopyranosyl-(1->4)-alpha-D-glucopyranose at 1.9 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Mir, R, Vikram, G, Sinha, M, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2009-09-25 | | Release date: | 2009-10-13 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Specific interactions of C-terminal half (C-lobe) of lactoferrin protein with edible sugars: binding and structural studies with implications on diabetes.
Int.J.Biol.Macromol., 47, 2010
|
|
3KK0
 
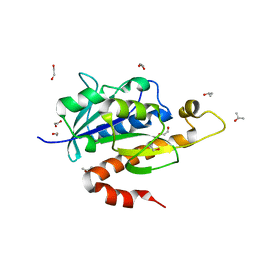 | | Crystal structure of partially folded intermediate state of peptidyl-tRNA hydrolase from Mycobacterium smegmatis | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kumar, A, Singh, N, Yadav, R, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2009-11-04 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of Fully-Folded Native and Partially-Folded Intermediate States of Peptidyl-tRNA Hydrolase from Mycobacterium smegmatis
To be Published
|
|
4YEH
 
 | | Crystal structure of Mg2+ ion containing hemopexin fold from Kabuli chana (chickpea white) at 2.45A resolution reveals a structural basis of metal ion transport | | Descriptor: | Lectin, MAGNESIUM ION | | Authors: | Kumar, S, Singh, A, Yamini, S, Bhushan, A, Dey, S, Sharma, S, Singh, T.P. | | Deposit date: | 2015-02-24 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Mg(2+) Containing Hemopexin-Fold Protein from Kabuli Chana (Chickpea-White, CW-25) at 2.45 angstrom Resolution Reveals Its Metal Ion Transport Property
Protein J., 34, 2015
|
|
4DHW
 
 | | Crystal structure of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa with Adipic acid at 2.4 Angstrom resolution | | Descriptor: | Peptidyl-tRNA hydrolase, hexanedioic acid | | Authors: | Kumar, A, Singh, A, Singh, N, Sinha, M, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2012-01-30 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal structure of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa with Adipic acid at 2.4 Angstrom resolution
To be Published
|
|
1PO8
 
 | | Crystal structure of a complex formed between krait venom phospholipase A2 and heptanoic acid at 2.7 A resolution. | | Descriptor: | HEPTANOIC ACID, Phospholipase A2, SODIUM ION | | Authors: | Singh, G, Jayasankar, J, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2003-06-14 | | Release date: | 2004-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of a complex formed between krait venom phospholipase A2 and heptanoic acid at 2.7 A resolution.
To be Published
|
|
1Q7A
 
 | | Crystal structure of the complex formed between russell's viper phospholipase A2 and an antiinflammatory agent oxyphenbutazone at 1.6A resolution | | Descriptor: | 4-BUTYL-1-(4-HYDROXYPHENYL)-2-PHENYLPYRAZOLIDINE-3,5-DIONE, METHANOL, Phospholipase A2 VRV-PL-VIIIa, ... | | Authors: | Singh, N, Jabeen, T, Sharma, S, Singh, T.P. | | Deposit date: | 2003-08-17 | | Release date: | 2004-05-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Phospholipase A2 as a target protein for nonsteroidal anti-inflammatory drugs (NSAIDS): crystal structure of the complex formed between phospholipase A2 and oxyphenbutazone at 1.6 A resolution.
Biochemistry, 43, 2004
|
|
4MTV
 
 | | Crystal structure of the complex of Buffalo Signalling Glycoprotein with pentasaccharide at 2.8A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1 | | Authors: | Shukla, P.K, Chaudhary, A, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-09-20 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the complex of Buffalo Signalling Glycoprotein with pentasaccharide at 2.8A resolution
To be Published
|
|
3SXV
 
 | | Crystal structure of the complex of goat lactoperoxidase with amitrole at 2.1 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pandey, N, Singh, R.P, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-07-15 | | Release date: | 2011-08-17 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the complex of goat lactoperoxidase with amitrole at 2.1 A resolution
TO BE PUBLISHED
|
|
5GH0
 
 | | Crystal structure of the complex of bovine lactoperoxidase with mercaptoimidazole at 2.3 A resolution | | Descriptor: | 1,3-dihydroimidazole-2-thione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, P.K, Sirohi, H.V, Singh, A.K, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-06-17 | | Release date: | 2016-06-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of anti-thyroid drugs: Binding studies and structure determination of the complex of lactoperoxidase with 2-mercaptoimidazole at 2.30 angstrom resolution
Proteins, 85, 2017
|
|
3GCI
 
 | | Crystal Structure of the Complex Formed Between a New Isoform of Phospholipase A2 with C-terminal Amyloid Beta Heptapeptide at 2 A Resolution | | Descriptor: | CALCIUM ION, Heptapeptide from Amyloid beta A4 protein, Phospholipase A2 isoform 3 | | Authors: | Mirza, Z, Vikram, G, Singh, N, Sinha, M, Bhushan, A, Sharma, S, Srinivasan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2009-02-22 | | Release date: | 2009-03-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal Structure of the Complex Formed Between a New Isoform of Phospholipase A2 with C-terminal Amyloid Beta Heptapeptide at 2 A Resolution
To be Published
|
|
3TGY
 
 | | Crystal structure of the complex of Bovine Lactoperoxidase with Ascorbic acid at 2.35 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamini, S, Singh, R.P, Singh, A.K, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-08-18 | | Release date: | 2011-09-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of bovine lactoperoxidase with a partially linked heme moiety at 1.98 angstrom resolution.
Biochim.Biophys.Acta, 1865, 2017
|
|
8Y9X
 
 | | Crystal structure of the complex of lactoperoxidase with four inorganic substrates, SCN, I, Br and Cl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, CALCIUM ION, ... | | Authors: | Viswanathan, V, Singh, A.K, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2024-02-07 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for the order of preference of inorganic substrates in mammalian heme peroxidases: crystal structure of the complex of lactoperoxidase with four inorganic substrates, SCN, I, Br and Cl
To Be Published
|
|
8ING
 
 | | Structure of the ternary complex of lactoperoxidase with substrate nitric oxide (NO) and product nitrite ion (NO2) at 1.98 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ahmad, M.I, Viswanathan, V, Kumar, M, Singh, R.P, Singh, A.K, Sinha, M, Kaur, P, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2023-03-09 | | Release date: | 2023-04-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of the ternary complex of lactoperoxidase with substrate nitric oxide (NO) and product nitrite ion (NO2) at 1.98 A resolution
To be published
|
|
1ZM6
 
 | | Crystal structure of the complex formed beween a group I phospholipase A2 and designed penta peptide Leu-Ala-Ile-Tyr-Ser at 2.6A resolution | | Descriptor: | ACETATE ION, Phospholipase A2 isoform 3, designed penta peptide Leu-Ala-Ile-Tyr-Ser | | Authors: | Singh, R.K, Singh, N, Jabeen, T, Sharma, S, Dey, S, Singh, T.P. | | Deposit date: | 2005-05-10 | | Release date: | 2005-06-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the complex of group I PLA2 with a group II-specific peptide Leu-Ala-Ile-Tyr-Ser (LAIYS) at 2.6 A resolution.
J.Drug Target., 13, 2005
|
|
1YXH
 
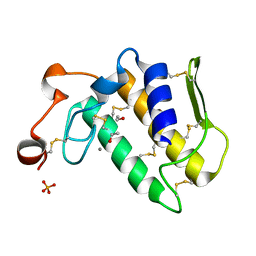 | | Crystal structure of a novel phospholipase A2 from Naja naja sagittifera with a strong anticoagulant activity | | Descriptor: | CALCIUM ION, ETHANOL, PHOSPHATE ION, ... | | Authors: | Jabeen, T, Singh, N, Singh, R.K, Sharma, S, Srinivasan, A, Singh, T.P. | | Deposit date: | 2005-02-21 | | Release date: | 2005-05-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of a novel phospholipase A(2) from Naja naja sagittifera with a strong anticoagulant activity
Toxicon, 46, 2005
|
|
5Y48
 
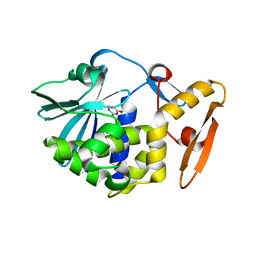 | | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina with Pyrimidine-2,4-dione at 1.70 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome inactivating protein, URACIL | | Authors: | Singh, P.K, Pandey, S, Iqbal, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-08-01 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein from Momordica balsamina with uracil and uridine.
Proteins, 87, 2019
|
|
4Y55
 
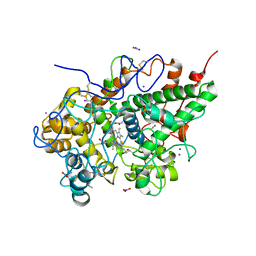 | | Crystal structure of Buffalo lactoperoxidase with Rhodanide at 2.09 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Gupta, A, Tyagi, T.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-02-11 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Buffalo lactoperoxidase with Rhodanide at 2.09 Angstrom resolution
To Be Published
|
|
4RHC
 
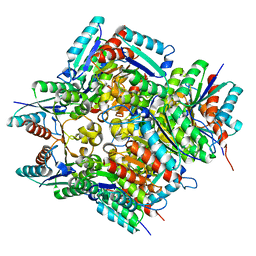 | |
5GLS
 
 | | Structure of bovine Lactoperoxidase with a partially modified covalent bond with heme moiety | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tiwari, P, Singh, P.K, Sirohi, H.V, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-07-12 | | Release date: | 2016-07-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of bovine lactoperoxidase with a partially linked heme moiety at 1.98 angstrom resolution
Biochim. Biophys. Acta, 1865, 2016
|
|
3TAK
 
 | | Crystal structure of the complex of DHDPS from Acinetobacter baumannii with Pyruvate at 1.4 A resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, Dihydrodipicolinate synthase, GLYCEROL, ... | | Authors: | Singh, A, Kaushik, S, Sinha, M, Tewari, R, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-08-04 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structure of the complex of DHDPS from Acinetobacter baumannii with Pyruvate at 1.4 A resolution
To be Published
|
|
9IIL
 
 | | Structure of the complex of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii with nicotinamide adenine dinucleotide in the presence of poly(ethylene glycol) at 2.20 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Jeyakanthan, J, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-06-20 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the complex of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii with nicotinamide adenine dinucleotide in the presence of poly(ethylene glycol) at 2.20 A resolution
To Be Published
|
|
9IIM
 
 | | Structure of the complex of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii with nicotinamide adenine dinucleotide at 2.74 A resolution. | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Jeyakanthan, J, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-06-20 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure of the complex of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii with nicotinamide adenine dinucleotide at 2.74 A resolution.
To Be Published
|
|
9IJ6
 
 | | Crystal structure of the complex of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii with Adenosine phosphate at 2.40 A resolution. | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glyceraldehyde-3-phosphate dehydrogenase, MAGNESIUM ION, ... | | Authors: | Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Jeyakanthan, J, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-06-21 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the complex of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii with Adenosine phosphate at 2.40 A resolution.
To Be Published
|
|
