4JST
 
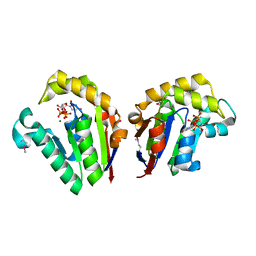 | | Structure of Clostridium thermocellum polynucleotide kinase bound to UTP | | Descriptor: | MAGNESIUM ION, Metallophosphoesterase, SODIUM ION, ... | | Authors: | Das, U, Wang, L.K, Smith, P, Shuman, S. | | Deposit date: | 2013-03-22 | | Release date: | 2013-08-28 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural and biochemical analysis of the phosphate donor specificity of the polynucleotide kinase component of the bacterial pnkphen1 RNA repair system.
Biochemistry, 52, 2013
|
|
6NVO
 
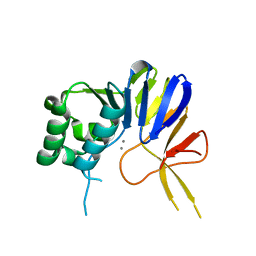 | | Crystal structure of Pseudomonas putida nuclease MPE | | Descriptor: | MANGANESE (II) ION, Nuclease MPE | | Authors: | Goldgur, Y, Shuman, S, Ejaz, A. | | Deposit date: | 2019-02-05 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Activity and structure ofPseudomonas putidaMPE, a manganese-dependent single-strand DNA endonuclease encoded in a nucleic acid repair gene cluster.
J.Biol.Chem., 294, 2019
|
|
4JT4
 
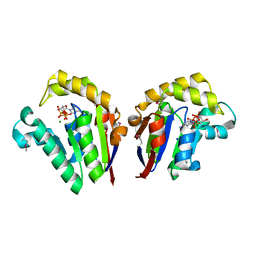 | | Structure of Clostridium thermocellum polynucleotide kinase bound to dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, MAGNESIUM ION, Metallophosphoesterase, ... | | Authors: | Das, U, Wang, L.K, Smith, P, Shuman, S. | | Deposit date: | 2013-03-22 | | Release date: | 2013-08-28 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and biochemical analysis of the phosphate donor specificity of the polynucleotide kinase component of the bacterial pnkphen1 RNA repair system.
Biochemistry, 52, 2013
|
|
4JT2
 
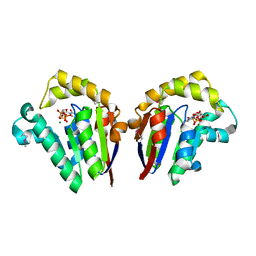 | | Structure of Clostridium thermocellum polynucleotide kinase bound to CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Metallophosphoesterase | | Authors: | Das, U, Wang, L.K, Smith, P, Shuman, S. | | Deposit date: | 2013-03-22 | | Release date: | 2013-08-28 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural and biochemical analysis of the phosphate donor specificity of the polynucleotide kinase component of the bacterial pnkphen1 RNA repair system.
Biochemistry, 52, 2013
|
|
7LFQ
 
 | | Pyrococcus RNA ligase | | Descriptor: | DNA (5'-D(*AP*TP*GP*TP*CP*C)-3'), POTASSIUM ION, RNA-splicing ligase RtcB, ... | | Authors: | Goldgur, Y, Shuman, S, Banerjee, A. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of 3'-PO 4 /5'-OH RNA ligase RtcB in complex with a 5'-OH oligonucleotide.
Rna, 27, 2021
|
|
4JSY
 
 | | Structure of Clostridium thermocellum polynucleotide kinase bound to GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Metallophosphoesterase | | Authors: | Das, U, Wang, L.K, Smith, P, Shuman, S. | | Deposit date: | 2013-03-22 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural and biochemical analysis of the phosphate donor specificity of the polynucleotide kinase component of the bacterial pnkphen1 RNA repair system.
Biochemistry, 52, 2013
|
|
1MQP
 
 | | THE CRYSTAL STRUCTURE OF ALPHA-D-GLUCURONIDASE FROM BACILLUS STEAROTHERMOPHILUS T-6 | | Descriptor: | GLYCEROL, alpha-D-glucuronidase | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2002-09-17 | | Release date: | 2003-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of Geobacillus stearothermophilus alpha-glucuronidase complexed with its substrate and products: mechanistic implications.
J.Biol.Chem., 279, 2004
|
|
2LJ6
 
 | |
1P16
 
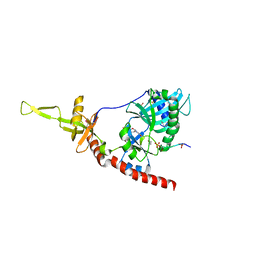 | | Structure of an mRNA capping enzyme bound to the phosphorylated carboxyl-terminal domain of RNA polymerase II | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Fabrega, C, Shen, V, Shuman, S, Lima, C.D. | | Deposit date: | 2003-04-11 | | Release date: | 2003-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of an mRNA capping enzyme bound to the phosphorylated carboxy-terminal domain of RNA polymerase II.
Mol.Cell, 11, 2003
|
|
2Q2T
 
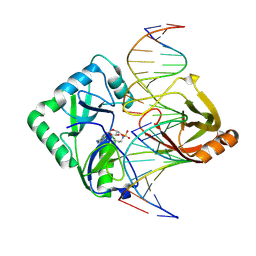 | | Structure of Chlorella virus DNA ligase-adenylate bound to a 5' phosphorylated nick | | Descriptor: | 5'-D(*AP*TP*TP*GP*CP*GP*AP*CP*(OMC)P*C)-3', 5'-D(*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*T)-3', 5'-D(P*CP*AP*CP*TP*AP*TP*CP*GP*GP*AP*A)-3', ... | | Authors: | Lima, C.D, Nandakumar, J, Nair, P.A, Smith, P, Shuman, S. | | Deposit date: | 2007-05-29 | | Release date: | 2007-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for nick recognition by a minimal pluripotent DNA ligase.
Nat.Struct.Mol.Biol., 14, 2007
|
|
9D8A
 
 | |
5COV
 
 | |
1I9S
 
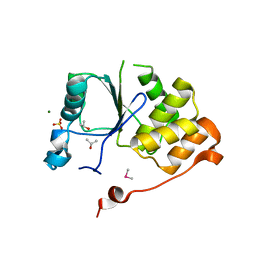 | | CRYSTAL STRUCTURE OF THE RNA TRIPHOSPHATASE DOMAIN OF MOUSE MRNA CAPPING ENZYME | | Descriptor: | CACODYLATE ION, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Changela, A, Ho, C.K, Martins, A, Shuman, S, Mondragon, A. | | Deposit date: | 2001-03-20 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and mechanism of the RNA triphosphatase component of mammalian mRNA capping enzyme.
EMBO J., 20, 2001
|
|
1LY1
 
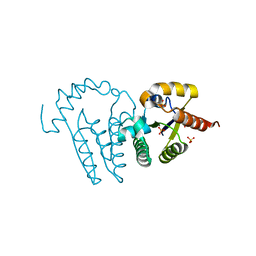 | |
5COT
 
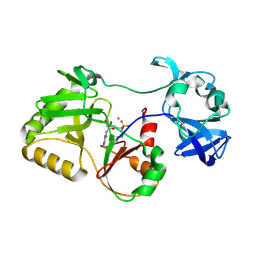 | |
5COU
 
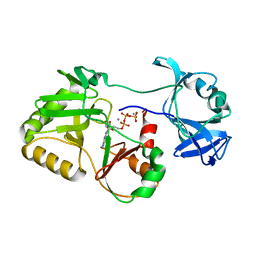 | |
6VT4
 
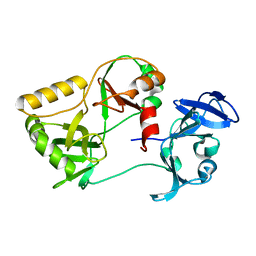 | | Naegleria gruberi RNA ligase R149A mutant apo | | Descriptor: | RNA Ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VTE
 
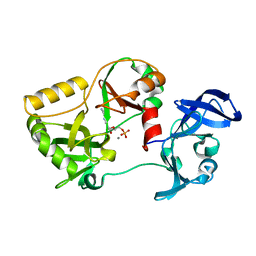 | | Naegleria gruberi RNA Ligase K170M mutant with AMP and Mn | | Descriptor: | ADENOSINE MONOPHOSPHATE, MANGANESE (II) ION, RNA ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VT8
 
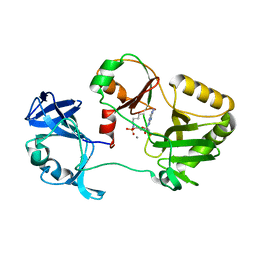 | | Naegleria gruberi RNA ligase E312A mutant with AMP and Mn | | Descriptor: | ADENOSINE MONOPHOSPHATE, MANGANESE (II) ION, RNA Ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VTD
 
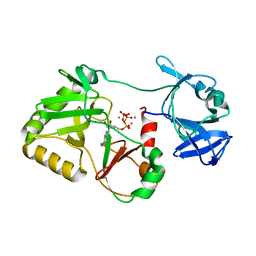 | | Naegleria gruberi RNA ligase R149A mutant with ATP and Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, RNA Ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VT6
 
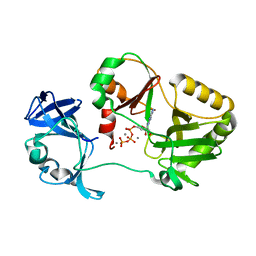 | | Naegleria gruberi RNA ligase K170A mutant with ATP and Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, RNA Ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VTG
 
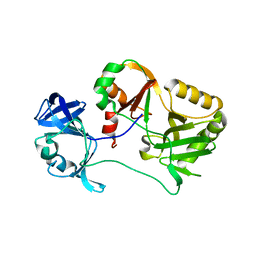 | | Naegleria gruberi RNA ligase E227A mutant apo | | Descriptor: | RNA ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VT9
 
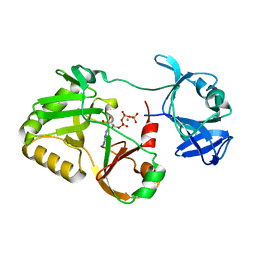 | | Naegleria gruberi RNA ligase E227A mutant with ATP and Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, RNA ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VT0
 
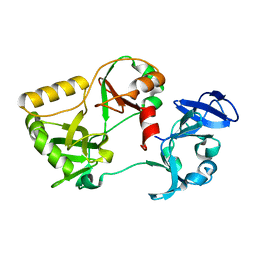 | | Naegleria gruberi RNA ligase K170A mutant apo | | Descriptor: | RNA Ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VTB
 
 | | Naegleria gruberi RNA ligase K326A mutant with ATP and Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, RNA ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.547 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
