1OVK
 
 | | T4 Lysozyme Cavity Mutant L99A/M102Q Bound with N-Allyl-Aniline | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme, ... | | Authors: | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | Deposit date: | 2003-03-26 | | Release date: | 2004-04-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Testing a Flexible-receptor Docking Algorithm in a Model Binding Site
J.Mol.Biol., 337, 2004
|
|
1OVH
 
 | | T4 Lysozyme Cavity Mutant L99A/M102Q Bound With 2-Chloro-6-Methyl-Aniline | | Descriptor: | 2-CHLORO-6-METHYL-ANILINE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | Deposit date: | 2003-03-26 | | Release date: | 2004-04-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Testing a Flexible-receptor Docking Algorithm in a Model Binding Site
J.Mol.Biol., 337, 2004
|
|
1OWY
 
 | | T4 Lysozyme Cavity Mutant L99A/M102Q Bound With 2-Propyl-Aniline | | Descriptor: | 2-PROPYL-ANILINE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | Deposit date: | 2003-03-31 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Testing a Flexible-receptor Docking Algorithm in a Model Binding Site
J.Mol.Biol., 337, 2004
|
|
1OV5
 
 | | T4 Lysozyme Cavity Mutant L99a/M102Q Bound With 2-Allylphenol | | Descriptor: | 2-ALLYLPHENOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | Deposit date: | 2003-03-25 | | Release date: | 2004-04-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Testing a Flexible-receptor Docking Algorithm in a Model Binding Site
J.Mol.Biol., 337, 2004
|
|
1OV7
 
 | | T4 Lysozyme Cavity Mutant L99A/M102Q Bound with 2-Allyl-6-Methyl-Phenol | | Descriptor: | 2-ALLYL-6-METHYL-PHENOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | Deposit date: | 2003-03-25 | | Release date: | 2004-04-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Testing a Flexible-receptor Docking Algorithm in a Model Binding Site
J.Mol.Biol., 337, 2004
|
|
1OVJ
 
 | | T4 Lysozyme Cavity Mutant L99A/M102Q Bound with 3-Fluoro-2-Methyl_Aniline | | Descriptor: | 3-FLUORO-2-METHYL-ANILINE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | Deposit date: | 2003-03-26 | | Release date: | 2004-04-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Testing a Flexible-receptor Docking Algorithm in a Model Binding Site
J.Mol.Biol., 337, 2004
|
|
5JWV
 
 | | T4 Lysozyme L99A/M102Q with Ethylbenzene Bound | | Descriptor: | Endolysin, PHENYLETHANE | | Authors: | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | Deposit date: | 2016-05-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
5JWT
 
 | | T4 Lysozyme L99A/M102Q with Benzene Bound | | Descriptor: | BENZENE, Endolysin | | Authors: | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | Deposit date: | 2016-05-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
5JWU
 
 | | T4 Lysozyme L99A/M102Q with 1,2-Dihydro-1,2-azaborine Bound | | Descriptor: | 1,2-dihydro-1,2-azaborinine, CHLORIDE ION, Endolysin | | Authors: | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | Deposit date: | 2016-05-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
5JWS
 
 | | T4 Lysozyme L99A with 1-Hydro-2-ethyl-1,2-azaborine Bound | | Descriptor: | 2-ethyl-1,2-dihydro-1,2-azaborinine, CHLORIDE ION, Endolysin | | Authors: | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | Deposit date: | 2016-05-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
5JWW
 
 | | T4 Lysozyme L99A/M102Q with 1-Hydro-2-ethyl-1,2-azaborine Bound | | Descriptor: | 2-ethyl-1,2-dihydro-1,2-azaborinine, CHLORIDE ION, Endolysin | | Authors: | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | Deposit date: | 2016-05-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
7W7E
 
 | | Cryo-EM structure of the alpha2A adrenergic receptor GoA signaling complex bound to a biased agonist | | Descriptor: | 5-(3-bicyclo[4.2.0]octa-1,3,5-trienyl)-1,2,3,6-tetrahydropyridine, Alpha-2A adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, J, Fink, E.A, Shoichet, B.K, Du, Y. | | Deposit date: | 2021-12-04 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure-based discovery of nonopioid analgesics acting through the alpha 2A -adrenergic receptor.
Science, 377, 2022
|
|
7W6P
 
 | | Cryo-EM structure of the alpha2A adrenergic receptor GoA signaling complex bound to a G protein biased agonist | | Descriptor: | Alpha-2A adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, J, Fink, E.A, Shoichet, B.K, Du, Y. | | Deposit date: | 2021-12-02 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structure-based discovery of nonopioid analgesics acting through the alpha 2A -adrenergic receptor.
Science, 377, 2022
|
|
8DIB
 
 | |
8DIF
 
 | |
8DIG
 
 | |
8DIC
 
 | |
8DIE
 
 | |
8DII
 
 | |
8DIH
 
 | | Virtual screening for novel SARS-CoV-2 main protease non-covalent and covalent inhibitors | | Descriptor: | (1P,1'R)-1-(isoquinolin-4-yl)-2',3'-dihydrospiro[imidazolidine-4,1'-indene]-2,5-dione, 3C-like proteinase nsp5 | | Authors: | Singh, I, Shoichet, B.K. | | Deposit date: | 2022-06-29 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Large library docking for novel SARS-CoV-2 main protease non-covalent and covalent inhibitors.
Protein Sci., 32, 2023
|
|
8DID
 
 | |
7TXT
 
 | | Structure of human serotonin transporter bound to small molecule '8090 in lipid nanodisc and NaCl | | Descriptor: | 1-[4-(4-fluorophenyl)-1,3-thiazol-2-yl]piperazine, 15B8 Fab heavy chain, 15B8 Fab light chain, ... | | Authors: | Singh, I, Seth, A, Billesboelle, C.B, Braz, J, Rodriguiz, R.M, Roy, K, Bekele, B, Craik, V, Huang, X.P, Boytsov, D, Lak, P, O'Donnell, H, Sandtner, W, Roth, B.L, Basbaum, A.I, Wetsel, W.C, Manglik, A, Shoichet, B.K, Rudnick, G. | | Deposit date: | 2022-02-09 | | Release date: | 2023-03-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-based discovery of conformationally selective inhibitors of the serotonin transporter.
Cell, 186, 2023
|
|
1NYY
 
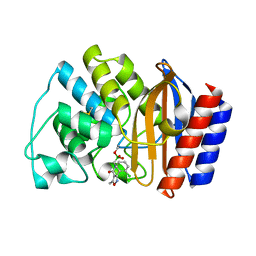 | | Crystal Structure of the complex between M182T mutant of TEM-1 and a boronic acid inhibitor (105) | | Descriptor: | Beta-lactamase TEM, N-[5-METHYL-3-O-TOLYL-ISOXAZOLE-4-CARBOXYLIC ACID AMIDE] BORONIC ACID | | Authors: | Wang, X, Minasov, G, Blazquez, J, Caselli, E, Prati, F, Shoichet, B.K. | | Deposit date: | 2003-02-14 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Recognition and resistance in TEM beta-lactamase
Biochemistry, 42, 2003
|
|
1NY0
 
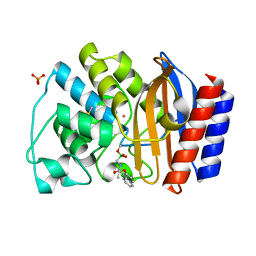 | | Crystal Structure of the complex between M182T mutant of TEM-1 and a boronic acid inhibitor (NBF) | | Descriptor: | Beta-lactamase TEM, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Wang, X, Minasov, G, Blazquez, J, Caselli, E, Prati, F, Shoichet, B.K. | | Deposit date: | 2003-02-11 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Recognition and Resistance in TEM beta-lactamase
Biochemistry, 42, 2003
|
|
1PI5
 
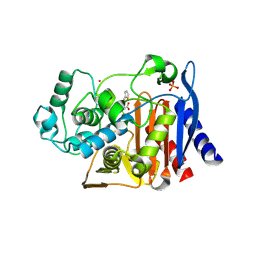 | | Structure of N289A mutant of AmpC in complex with SM2, carboxyphenylglycylboronic acid bearing the cephalothin R1 side chain | | Descriptor: | (1R)-1-(2-THIENYLACETYLAMINO)-1-(3-CARBOXYPHENYL)METHYLBORONIC ACID, Beta-lactamase, PHOSPHATE ION, ... | | Authors: | Roth, T.A, Minasov, G, Focia, P.J, Shoichet, B.K. | | Deposit date: | 2003-05-29 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Thermodynamic cycle analysis and inhibitor design against beta-lactamase.
Biochemistry, 42, 2003
|
|
