7DA2
 
 | | The crystal structure of the chicken FANCM-MHF complex | | Descriptor: | Centromere protein S, Centromere protein X, Fanconi anemia group M protein | | Authors: | Nishino, T, Ito, S. | | Deposit date: | 2020-10-14 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural analysis of the chicken FANCM-MHF complex and its stability.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
4L60
 
 | |
4L6P
 
 | |
2ZXE
 
 | | Crystal structure of the sodium - potassium pump in the E2.2K+.Pi state | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Shinoda, T, Ogawa, H, Cornelius, F, Toyoshima, C. | | Deposit date: | 2008-12-22 | | Release date: | 2009-05-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the sodium - potassium pump at 2.4 A resolution
Nature, 459, 2009
|
|
2D42
 
 | | Crystal structure analysis of a non-toxic crystal protein from Bacillus thuringiensis | | Descriptor: | non-toxic crystal protein | | Authors: | Akiba, T, Higuchi, K, Mizuki, E, Ekino, K, Shin, T, Ohba, M, Kanai, R, Harata, K. | | Deposit date: | 2005-10-05 | | Release date: | 2006-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Nontoxic crystal protein from Bacillus thuringiensis demonstrates a remarkable structural similarity to beta-pore-forming toxins
Proteins, 63, 2006
|
|
1PKT
 
 | | STRUCTURE OF THE PI3K SH3 DOMAIN AND ANALYSIS OF THE SH3 FAMILY | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT SH3 DOMAIN | | Authors: | Koyama, S, Yu, H, Dalgarno, D.C, Shin, T.B, Zydowsky, L.D, Schreiber, S.L. | | Deposit date: | 1994-03-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the PI3K SH3 domain and analysis of the SH3 family.
Cell(Cambridge,Mass.), 72, 1993
|
|
1PKS
 
 | | STRUCTURE OF THE PI3K SH3 DOMAIN AND ANALYSIS OF THE SH3 FAMILY | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT SH3 DOMAIN | | Authors: | Koyama, S, Yu, H, Dalgarno, D.C, Shin, T.B, Zydowsky, L.D, Schreiber, S.L. | | Deposit date: | 1994-03-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the PI3K SH3 domain and analysis of the SH3 family.
Cell(Cambridge,Mass.), 72, 1993
|
|
1LM4
 
 | | Structure of Peptide Deformylase from Staphylococcus aureus at 1.45 A | | Descriptor: | FE (III) ION, GLYCEROL, peptide deformylase PDF1 | | Authors: | Kreusch, A, Spraggon, G, Lee, C.C, Klock, H, McMullan, D, Ng, K, Shin, T, Vincent, J, Warner, I, Ericson, C, Lesley, S.A. | | Deposit date: | 2002-04-30 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure analysis of peptide deformylases from streptococcus pneumoniae,staphylococcus aureus, thermotoga maritima, and pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase
J.MOL.BIOL., 330, 2003
|
|
1LME
 
 | | Crystal Structure of Peptide Deformylase from Thermotoga maritima | | Descriptor: | peptide deformylase | | Authors: | Kreusch, A, Spraggon, G, Lee, C.C, Klock, H, McMullan, D, Ng, K, Shin, T, Vincent, J, Warner, I, Ericson, C, Lesley, S.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2002-05-01 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure analysis of peptide deformylases from streptococcus pneumoniae,staphylococcus aureus, thermotoga maritima, and pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase
J.MOL.BIOL., 330, 2003
|
|
1LM6
 
 | | Crystal Structure of Peptide Deformylase from Streptococcus pneumoniae | | Descriptor: | FE (III) ION, GLYCEROL, peptide deformylase DEFB | | Authors: | Kreusch, A, Spraggon, G, Lee, C.C, Klock, H, McMullan, D, Ng, K, Shin, T, Vincent, J, Warner, I, Ericson, C, Lesley, S.A. | | Deposit date: | 2002-04-30 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure analysis of peptide deformylases from streptococcus pneumoniae,staphylococcus aureus, thermotoga maritima, and pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase
J.MOL.BIOL., 330, 2003
|
|
1N5N
 
 | | Crystal Structure of Peptide Deformylase from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, Peptide deformylase, ZINC ION | | Authors: | Kreusch, A, Spraggon, G, Lee, C.C, Klock, H, McMullan, D, Ng, K, Shin, T, Vincent, J, Warner, I, Ericson, C, Lesley, S.A. | | Deposit date: | 2002-11-06 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure analysis of peptide deformylases from streptococcus pneumoniae,staphylococcus aureus, thermotoga maritima, and pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase
J.MOL.BIOL., 330, 2003
|
|
1SRM
 
 | | 1H AND 15N ASSIGNMENTS AND SECONDARY STRUCTURE OF THE SRC SH3 DOMAIN | | Descriptor: | SRC TYROSINE KINASE SH3 DOMAIN | | Authors: | Yu, H, Rosen, M.K, Shin, T.B, Seidel-Dugan, C, Brugge, J.S, Schreiber, S.L. | | Deposit date: | 1994-03-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | 1H and 15N assignments and secondary structure of the Src SH3 domain.
FEBS Lett., 324, 1993
|
|
1SRL
 
 | | 1H AND 15N ASSIGNMENTS AND SECONDARY STRUCTURE OF THE SRC SH3 DOMAIN | | Descriptor: | SRC TYROSINE KINASE SH3 DOMAIN | | Authors: | Yu, H, Rosen, M.K, Shin, T.B, Seidel-Dugan, C, Brugge, J.S, Schreiber, S.L. | | Deposit date: | 1994-03-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | 1H and 15N assignments and secondary structure of the Src SH3 domain.
FEBS Lett., 324, 1993
|
|
3NLA
 
 | | NMR STRUCTURE OF THE N-TERMINAL DOMAIN WITH A LINKER PORTION OF ANTARCTIC EEL POUT ANTIFREEZE PROTEIN RD3, 40 STRUCTURES | | Descriptor: | ANTIFREEZE PROTEIN RD3 TYPE III | | Authors: | Miura, K, Ohgiya, S, Hoshino, T, Nemoto, N, Hikichi, K, Tsuda, S. | | Deposit date: | 1998-02-24 | | Release date: | 1999-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of a globular antifreeze protein to ice.
Nature, 384, 1996
|
|
1FO4
 
 | | CRYSTAL STRUCTURE OF XANTHINE DEHYDROGENASE ISOLATED FROM BOVINE MILK | | Descriptor: | 2-HYDROXYBENZOIC ACID, CALCIUM ION, DIOXOTHIOMOLYBDENUM(VI) ION, ... | | Authors: | Enroth, C, Eger, B.T, Okamoto, K, Nishino, T, Nishino, T, Pai, E.F. | | Deposit date: | 2000-08-24 | | Release date: | 2000-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of bovine milk xanthine dehydrogenase and xanthine oxidase: structure-based mechanism of conversion.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1FIQ
 
 | | CRYSTAL STRUCTURE OF XANTHINE OXIDASE FROM BOVINE MILK | | Descriptor: | 2-HYDROXYBENZOIC ACID, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Enroth, C, Eger, B.T, Okamoto, K, Nishino, T, Nishino, T, Pai, E.F. | | Deposit date: | 2000-08-04 | | Release date: | 2000-10-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of bovine milk xanthine dehydrogenase and xanthine oxidase: structure-based mechanism of conversion.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
3BDJ
 
 | | Crystal Structure of Bovine Milk Xanthine Dehydrogenase with a Covalently Bound Oxipurinol Inhibitor | | Descriptor: | CALCIUM ION, CARBONATE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Eger, B.T, Okamoto, K, Nishino, T, Pai, E.F, Nishino, T. | | Deposit date: | 2007-11-14 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of inhibition of xanthine oxidoreductase by allopurinol: crystal structure of reduced bovine milk xanthine oxidoreductase bound with oxipurinol.
Nucleosides Nucleotides Nucleic Acids, 27, 2008
|
|
3AMZ
 
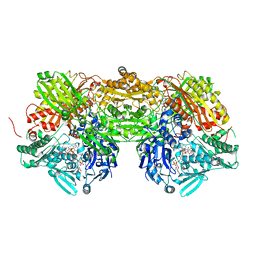 | | Bovine Xanthine Oxidoreductase urate bound form | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, BICARBONATE ION, CALCIUM ION, ... | | Authors: | Okamoto, K, Eger, B.T, Pai, E.F, Nishino, T. | | Deposit date: | 2010-08-27 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Urate Bound Form of Xanthine Oxidoreductase: Substrate Orientation and Structure of the Key Reaction Intermediate
J.Am.Chem.Soc., 132, 2010
|
|
3AM9
 
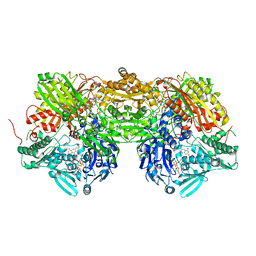 | | Complex of bovine xanthine dehydrogenase and trihydroxy FYX-051 | | Descriptor: | 4-[5-(2,6-dioxo-1,2,3,6-tetrahydropyridin-4-yl)-1H-1,2,4-triazol-3-yl]-6-oxo-1,6-dihydropyridine-2-carbonitrile, BICARBONATE ION, CALCIUM ION, ... | | Authors: | Matsumoto, K, Okamoto, K, Ashizawa, N, Matsumura, T, Kusano, T, Nishino, T. | | Deposit date: | 2010-08-18 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | FYX-051: A Novel and Potent Hybrid-Type Inhibitor of Xanthine Oxidoreductase
J.Pharmacol.Exp.Ther., 336, 2011
|
|
3AN1
 
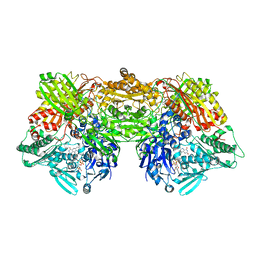 | | Crystal structure of rat D428A mutant, urate bound form | | Descriptor: | BICARBONATE ION, CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Okamoto, K, Kawaguchi, Y, Eger, B.T, Pai, E.F, Nishino, T. | | Deposit date: | 2010-08-27 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structures of Urate Bound Form of Xanthine Oxidoreductase: Substrate Orientation and Structure of the Key Reaction Intermediate
J.Am.Chem.Soc., 132, 2010
|
|
2E1Q
 
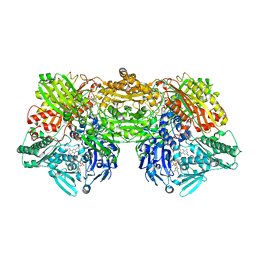 | | Crystal Structure of Human Xanthine Oxidoreductase mutant, Glu803Val | | Descriptor: | 2-HYDROXYBENZOIC ACID, BICARBONATE ION, CALCIUM ION, ... | | Authors: | Yamaguchi, Y, Matsumura, T, Ichida, K, Okamoto, K, Nishino, T. | | Deposit date: | 2006-10-27 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human xanthine oxidase changes its substrate specificity to aldehyde oxidase type upon mutation of amino acid residues in the active site: roles of active site residues in binding and activation of purine substrate
J.Biochem.(Tokyo), 141, 2007
|
|
1C7Y
 
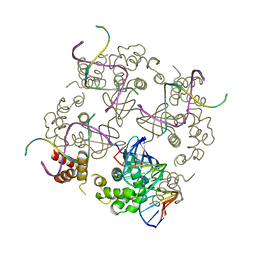 | | E.COLI RUVA-HOLLIDAY JUNCTION COMPLEX | | Descriptor: | DNA (5'-D(P*DAP*DAP*DGP*DTP*DTP*DGP*DGP*DGP*DAP*DTP*DTP*DGP*DT)-3'), DNA (5'-D(P*DCP*DAP*DAP*DTP*DCP*DCP*DCP*DAP*DAP*DCP*DTP*DT)-3'), DNA (5'-D(P*DCP*DGP*DAP*DAP*DTP*DGP*DTP*DGP*DTP*DGP*DTP*DCP*DT)-3'), ... | | Authors: | Ariyoshi, M, Nishino, T, Iwasaki, H, Shinagawa, H, Morikawa, K. | | Deposit date: | 2000-04-03 | | Release date: | 2000-07-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the holliday junction DNA in complex with a single RuvA tetramer.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1N5X
 
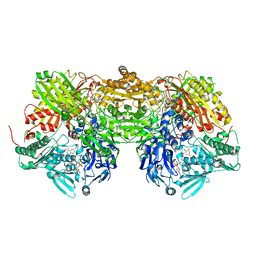 | | Xanthine Dehydrogenase from Bovine Milk with Inhibitor TEI-6720 Bound | | Descriptor: | 2-(3-CYANO-4-ISOBUTOXY-PHENYL)-4-METHYL-5-THIAZOLE-CARBOXYLIC ACID, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Okamoto, K, Eger, B.T, Nishino, T, Kondo, S, Pai, E.F, Nishino, T. | | Deposit date: | 2002-11-07 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An Extremely Potent Inhibitor of Xanthine Oxidoreductase: Crystal Structure of the Enzyme-Inhibitor Complex and Mechanism of Inhibition
J.BIOL.CHEM., 278, 2003
|
|
2E3T
 
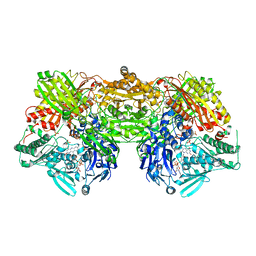 | | Crystal structure of rat xanthine oxidoreductase mutant (W335A and F336L) | | Descriptor: | BICARBONATE ION, CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Asai, R, Nishino, T, Matsumura, T, Okamoto, K, Pai, E.F, Nishino, T. | | Deposit date: | 2006-11-28 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Two mutations convert mammalian xanthine oxidoreductase to highly superoxide-productive xanthine oxidase
J.Biochem.(Tokyo), 141, 2007
|
|
8Z5L
 
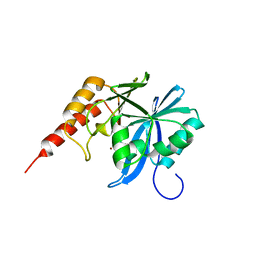 | | Crystal structure of metallo-beta-lactamse, IMP-1, complexed with a quinolinone-based inhibitor | | Descriptor: | 3-[2-azanyl-5-[2-cyclohexylethyl-[3-(4-methylphenoxy)propyl]amino]phenyl]propanoic acid, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Kamo, T, Kuroda, K, Nimura, S, Guo, Y, Kondo, S, Nukaga, M, Hoshino, T. | | Deposit date: | 2024-04-18 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Development of Inhibitory Compounds for Metallo-beta-lactamase through Computational Design and Crystallographic Analysis.
Biochemistry, 63, 2024
|
|
