7XJ7
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | Descriptor: | MANGANESE (II) ION, Reverse Transcriptase RNase H domain, ZINC ION, ... | | Authors: | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | Deposit date: | 2022-04-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
7XJ4
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | Descriptor: | MANGANESE (II) ION, Reverse Transcriptase RNase H domain, S-[5-[(E)-2-phenylethenyl]-1,3,4-oxadiazol-2-yl] 5-nitrothiophene-2-carbothioate, ... | | Authors: | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | Deposit date: | 2022-04-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
7XIS
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | Descriptor: | (2-methoxy-4-methoxycarbonyl-phenyl) 5-nitrofuran-2-carboxylate, MANGANESE (II) ION, Reverse Transcriptase RNase H domain, ... | | Authors: | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | Deposit date: | 2022-04-14 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
2YX8
 
 | | Crystal structure of the extracellular domain of human RAMP1 | | Descriptor: | Receptor activity-modifying protein 1 | | Authors: | Kusano, S, Kukimoto-Niino, M, Shirouzu, M, Shindo, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-24 | | Release date: | 2008-04-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human receptor activity-modifying protein 1 extracellular domain.
Protein Sci., 17, 2008
|
|
7DDM
 
 | | Crystal Structure of PenA39 beta-Lactamase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ALA-ALA-ARG-ASP-ALA-ALA-VAL-SER-ASP-ALA-ALA-ALA, ... | | Authors: | Nukaga, M, Hoshino, T, Papp-Wallace, K.M. | | Deposit date: | 2020-10-29 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Frameshift mutations in genes encoding PBP3 and PBP4 trigger an unusual, extreme beta-lactam resistance phenotype in Burkholderia multivorans
To Be Published
|
|
1SN7
 
 | | KUMAMOLISIN-AS, APOENZYME | | Descriptor: | CALCIUM ION, kumamolisin-As | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Oda, K, Nishino, T. | | Deposit date: | 2004-03-10 | | Release date: | 2004-06-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and biochemical investigations of kumamolisin-as, a serine-carboxyl peptidase with collagenase activity.
J.Biol.Chem., 279, 2004
|
|
3PGE
 
 | | Structure of sumoylated PCNA | | Descriptor: | Proliferating cell nuclear antigen, SUMO-modified proliferating cell nuclear antigen | | Authors: | Freudenthal, B.D, Brogie, J.E, Gakhar, L, Washington, T. | | Deposit date: | 2010-11-01 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of SUMO-Modified Proliferating Cell Nuclear Antigen.
J.Mol.Biol., 406, 2011
|
|
1FIY
 
 | | THREE-DIMENSIONAL STRUCTURE OF PHOSPHOENOLPYRUVATE CARBOXYLASE FROM ESCHERICHIA COLI AT 2.8 A RESOLUTION | | Descriptor: | ASPARTIC ACID, PHOSPHOENOLPYRUVATE CARBOXYLASE | | Authors: | Kai, Y, Matsumura, H, Inoue, T, Terada, K, Nagara, Y, Yoshinaga, T, Kihara, A, Izui, K. | | Deposit date: | 1998-05-02 | | Release date: | 1999-02-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of phosphoenolpyruvate carboxylase: a proposed mechanism for allosteric inhibition.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3WQK
 
 | | Crystal structure of Rv3378c with PO4 | | Descriptor: | Diterpene synthase, PHOSPHATE ION | | Authors: | Chan, H.C, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Zheng, Y, Bogue, S, Nakano, C, Hoshino, T, Zhang, L, Lv, P, Liu, W, Crick, D.C, Liang, P.H, Wang, A.H, Oldfield, E, Guo, R.T. | | Deposit date: | 2014-01-28 | | Release date: | 2014-02-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and inhibition of tuberculosinol synthase and decaprenyl diphosphate synthase from Mycobacterium tuberculosis.
J.Am.Chem.Soc., 136, 2014
|
|
3WQM
 
 | | Crystal structure of Rv3378c with inhibitor BPH-629 | | Descriptor: | Diterpene synthase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Chan, H.C, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Zheng, Y, Bogue, S, Nakano, C, Hoshino, T, Zhang, L, Lv, P, Liu, W, Crick, D.C, Liang, P.H, Wang, A.H, Oldfield, E, Guo, R.T. | | Deposit date: | 2014-01-28 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and inhibition of tuberculosinol synthase and decaprenyl diphosphate synthase from Mycobacterium tuberculosis.
J.Am.Chem.Soc., 136, 2014
|
|
3WQN
 
 | | Crystal structure of Rv3378c_Y51F with TPP | | Descriptor: | (2E)-3-methyl-5-[(1R,2S,8aS)-1,2,5,5-tetramethyl-1,2,3,5,6,7,8,8a-octahydronaphthalen-1-yl]pent-2-en-1-yl trihydrogen diphosphate, Diterpene synthase, PHOSPHATE ION | | Authors: | Chan, H.C, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Zheng, Y, Bogue, S, Nakano, C, Hoshino, T, Zhang, L, Lv, P, Liu, W, Crick, D.C, Liang, P.H, Wang, A.H, Oldfield, E, Guo, R.T. | | Deposit date: | 2014-01-28 | | Release date: | 2014-02-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and inhibition of tuberculosinol synthase and decaprenyl diphosphate synthase from Mycobacterium tuberculosis.
J.Am.Chem.Soc., 136, 2014
|
|
3WQL
 
 | | Crystal structure of Rv3378c with Mg2+ and PPi | | Descriptor: | Diterpene synthase, MAGNESIUM ION | | Authors: | Chan, H.C, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Zheng, Y, Bogue, S, Nakano, C, Hoshino, T, Zhang, L, Lv, P, Liu, W, Crick, D.C, Liang, P.H, Wang, A.H, Oldfield, E, Guo, R.T. | | Deposit date: | 2014-01-28 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and inhibition of tuberculosinol synthase and decaprenyl diphosphate synthase from Mycobacterium tuberculosis.
J.Am.Chem.Soc., 136, 2014
|
|
3UNC
 
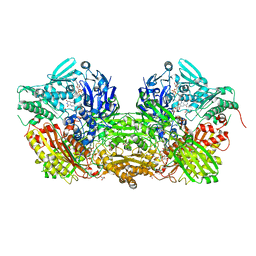 | | Crystal Structure of Bovine Milk Xanthine Dehydrogenase to 1.65A Resolution | | Descriptor: | 2-HYDROXYBENZOIC ACID, CALCIUM ION, CARBONATE ION, ... | | Authors: | Eger, B.T, Okamoto, K, Nishino, T, Pai, E.F. | | Deposit date: | 2011-11-15 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Protein conformational gating of enzymatic activity in xanthine oxidoreductase.
J.Am.Chem.Soc., 134, 2012
|
|
3UNI
 
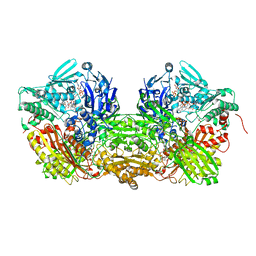 | | Crystal Structure of Bovine Milk Xanthine Dehydrogenase with NADH Bound | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-HYDROXYBENZOIC ACID, CALCIUM ION, ... | | Authors: | Eger, B.T, Okamoto, K, Nishino, T, Pai, E.F. | | Deposit date: | 2011-11-15 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein conformational gating of enzymatic activity in xanthine oxidoreductase.
J.Am.Chem.Soc., 134, 2012
|
|
3UNA
 
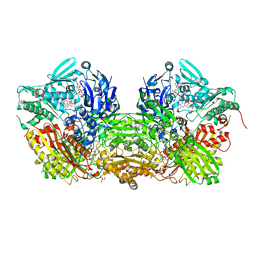 | | Crystal Structure of Bovine Milk Xanthine Dehydrogenase with NAD Bound | | Descriptor: | 2-HYDROXYBENZOIC ACID, CALCIUM ION, CARBONATE ION, ... | | Authors: | Eger, B.T, Okamoto, K, Nishino, T, Pai, E.F. | | Deposit date: | 2011-11-15 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein conformational gating of enzymatic activity in xanthine oxidoreductase.
J.Am.Chem.Soc., 134, 2012
|
|
2E1Q
 
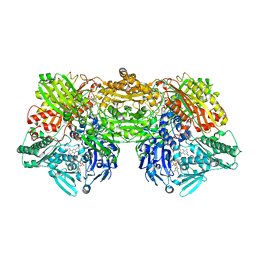 | | Crystal Structure of Human Xanthine Oxidoreductase mutant, Glu803Val | | Descriptor: | 2-HYDROXYBENZOIC ACID, BICARBONATE ION, CALCIUM ION, ... | | Authors: | Yamaguchi, Y, Matsumura, T, Ichida, K, Okamoto, K, Nishino, T. | | Deposit date: | 2006-10-27 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human xanthine oxidase changes its substrate specificity to aldehyde oxidase type upon mutation of amino acid residues in the active site: roles of active site residues in binding and activation of purine substrate
J.Biochem.(Tokyo), 141, 2007
|
|
7DA0
 
 | |
7DA1
 
 | | Crystal structure of the chicken MHF complex | | Descriptor: | Centromere protein S, Centromere protein X | | Authors: | Ito, S, Nishino, T. | | Deposit date: | 2020-10-14 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural analysis of the chicken FANCM-MHF complex and its stability.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7CJU
 
 | |
2FZA
 
 | | Crystal structure of d(GCGGGAGC): the base-intercalated duplex | | Descriptor: | 5'-D(*GP*(CBR)P*GP*GP*GP*AP*GP*C)-3', CALCIUM ION, SODIUM ION | | Authors: | Kondo, J, Ciengshin, T, Juan, E.C.M, Mitomi, K, Takenaka, A. | | Deposit date: | 2006-02-09 | | Release date: | 2007-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of d(gcGXGAgc) with X=G: a mutation at X is possible to occur in a base-intercalated duplex for multiplex formation
Nucleosides Nucleotides Nucleic Acids, 25, 2006
|
|
3WJ2
 
 | | Crystal structure of ESTFA (FE-lacking apo form) | | Descriptor: | Carboxylesterase | | Authors: | Ohara, K, Unno, H, Oshima, Y, Furukawa, K, Fujino, N, Hirooka, K, Hemmi, H, Takahashi, S, Nishino, T, Kusunoki, M, Nakayama, T. | | Deposit date: | 2013-10-03 | | Release date: | 2014-07-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural insights into the low pH adaptation of a unique carboxylesterase from Ferroplasma: altering the pH optima of two carboxylesterases.
J.Biol.Chem., 289, 2014
|
|
3WJ1
 
 | | Crystal structure of SSHESTI | | Descriptor: | Carboxylesterase, octyl beta-D-glucopyranoside | | Authors: | Ohara, K, Unno, H, Oshima, Y, Furukawa, K, Fujino, N, Hirooka, K, Hemmi, H, Takahashi, S, Nishino, T, Kusunoki, M, Nakayama, T. | | Deposit date: | 2013-10-03 | | Release date: | 2014-07-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the low pH adaptation of a unique carboxylesterase from Ferroplasma: altering the pH optima of two carboxylesterases.
J.Biol.Chem., 289, 2014
|
|
3A3Y
 
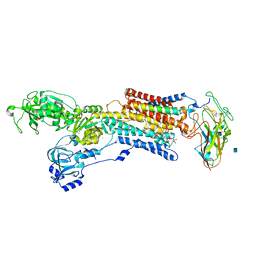 | | Crystal structure of the sodium-potassium pump with bound potassium and ouabain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, MAGNESIUM ION, ... | | Authors: | Ogawa, H, Shinoda, T, Cornelius, F, Toyoshima, C. | | Deposit date: | 2009-06-23 | | Release date: | 2009-09-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the sodium-potassium pump (Na+,K+-ATPase) with bound potassium and ouabain.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3AQF
 
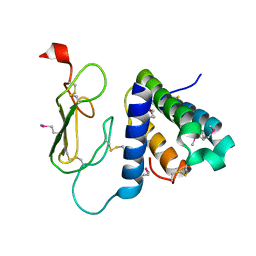 | | Crystal structure of the human CRLR/RAMP2 extracellular complex | | Descriptor: | Calcitonin gene-related peptide type 1 receptor, Receptor activity-modifying protein 2 | | Authors: | Kusano, S, Kukimono-Niino, M, Shirouzu, M, Shindo, T, Yokoyama, S. | | Deposit date: | 2010-10-29 | | Release date: | 2011-11-02 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for extracellular interactions between calcitonin receptor-like receptor and receptor activity-modifying protein 2 for adrenomedullin-specific binding
Protein Sci., 21, 2012
|
|
3AQE
 
 | | Crystal structure of the extracellular domain of human RAMP2 | | Descriptor: | Receptor activity-modifying protein 2 | | Authors: | Kusano, S, Kukimoto-Niino, M, Shirouzu, M, Shindo, T, Yokoyama, S. | | Deposit date: | 2010-10-29 | | Release date: | 2011-11-09 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for extracellular interactions between calcitonin receptor-like receptor and receptor activity-modifying protein 2 for adrenomedullin-specific binding
Protein Sci., 21, 2012
|
|
