2ZHS
 
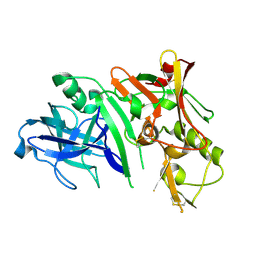 | | Crystal structure of BACE1 at pH 4.0 | | Descriptor: | Beta-secretase 1 | | Authors: | Shimizu, H, Nukina, N. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of an active form of BACE1, an enzyme responsible for amyloid beta protein production
Mol.Cell.Biol., 28, 2008
|
|
2ZHU
 
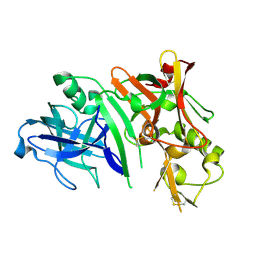 | | Crystal structure of BACE1 at pH 5.0 | | Descriptor: | Beta-secretase 1 | | Authors: | Shimizu, H, Nukina, N. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an active form of BACE1, an enzyme responsible for amyloid beta protein production
Mol.Cell.Biol., 28, 2008
|
|
2ZHT
 
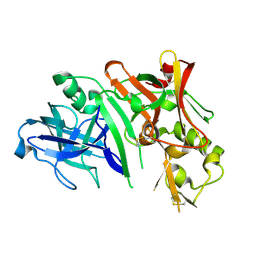 | | Crystal structure of BACE1 at pH 4.5 | | Descriptor: | Beta-secretase 1 | | Authors: | Shimizu, H, Nukina, N. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of an active form of BACE1, an enzyme responsible for amyloid beta protein production
Mol.Cell.Biol., 28, 2008
|
|
2ZHR
 
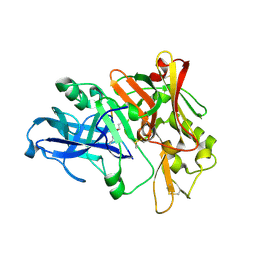 | |
3VRA
 
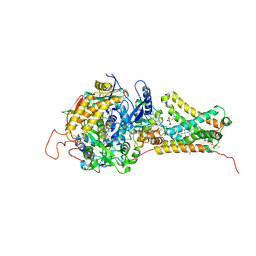 | | Mitochondrial rhodoquinol-fumarate reductase from the parasitic nematode Ascaris suum with the specific inhibitor Atpenin A5 | | Descriptor: | 3-[(2S,4S,5R)-5,6-DICHLORO-2,4-DIMETHYL-1-OXOHEXYL]-4-HYDROXY-5,6-DIMETHOXY-2(1H)-PYRIDINONE, Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Shimizu, H, Shiba, T, Inaoka, D.K, Osanai, A, Kita, K, Sakamoto, K, Harada, S. | | Deposit date: | 2012-04-07 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.44 Å) | | Cite: | Crystal structure of mitochondrial quinol-fumarate reductase from parasitic nematode Ascaris suum
To be Published
|
|
3VR9
 
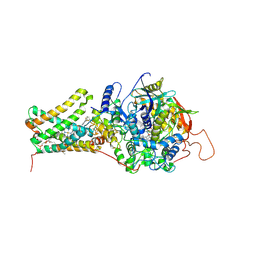 | | Mitochondrial rhodoquinol-fumarate reductase from the parasitic nematode Ascaris suum with the specific inhibitor flutolanil | | Descriptor: | Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Shimizu, H, Shiba, T, Inaoka, D.K, Osanai, A, Kita, K, Sakamoto, K, Harada, S. | | Deposit date: | 2012-04-07 | | Release date: | 2013-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal structure of mitochondrial quinol-fumarate reductase from parasitic nematode Ascaris suum
To be Published
|
|
3VR8
 
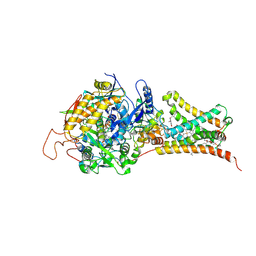 | | Mitochondrial rhodoquinol-fumarate reductase from the parasitic nematode Ascaris suum | | Descriptor: | 2-amino-3-methoxy-6-methyl-5-[(2E)-3-methylhex-2-en-1-yl]cyclohexa-2,5-diene-1,4-dione, Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Shimizu, H, Shiba, T, Inaoka, D.K, Osanai, A, Kita, K, Sakamoto, K, Harada, S. | | Deposit date: | 2012-04-07 | | Release date: | 2012-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structure of mitochondrial quinol-fumarate reductase from the parasitic nematode Ascaris suum
J.Biochem., 151, 2012
|
|
3VRB
 
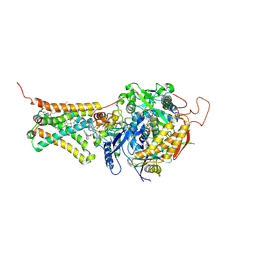 | | Mitochondrial rhodoquinol-fumarate reductase from the parasitic nematode Ascaris suum with the specific inhibitor flutolanil and substrate fumarate | | Descriptor: | Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Shimizu, H, Shiba, T, Inaoka, D.K, Osanai, A, Kita, K, Sakamoto, K, Harada, S. | | Deposit date: | 2012-04-07 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structure of mitochondrial quinol-fumarate reductase from the parasitic nematode Ascaris suum
J.Biochem., 151, 2012
|
|
3J3Z
 
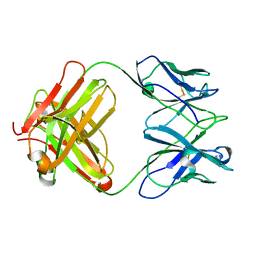 | | Structure of MA28-7 neutralizing antibody Fab fragment from electron cryo-microscopy of enterovirus 71 complexed with a Fab fragment | | Descriptor: | MA28-7 neutralizing antibody heavy chain, MA28-7 neutralizing antibody light chain | | Authors: | Lee, H, Cifuente, J.O, Ashley, R.E, Conway, J.F, Makhov, A.M, Tano, Y, Shimizu, H, Nishimura, Y, Hafenstein, S. | | Deposit date: | 2013-05-21 | | Release date: | 2013-08-28 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (23.4 Å) | | Cite: | A strain-specific epitope of enterovirus 71 identified by cryo-electron microscopy of the complex with fab from neutralizing antibody.
J.Virol., 87, 2013
|
|
7W86
 
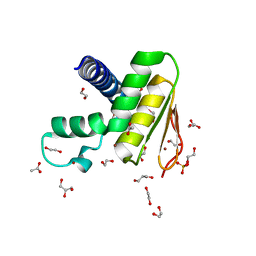 | | Crystal structure of the DYW domain of DYW1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Sawada, Y, Shimizu, H, Toma-Fukai, S, Shimizu, T. | | Deposit date: | 2021-12-07 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into the activation of an Arabidopsis organellar C-to-U RNA editing enzyme by active site complementation.
Plant Cell, 35, 2023
|
|
3Q2Z
 
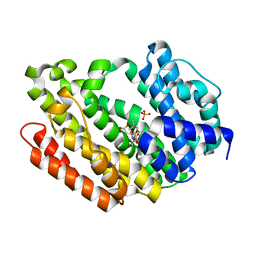 | | Human Squalene synthase in complex with N-[(3R,5S)-7-Chloro-5-(2,3-dimethoxyphenyl)-1-neopentyl-2-oxo-1,2,3,5-tetrahydro-4,1-benzoxazepine-3-acetyl]-L-aspartic acid | | Descriptor: | N-{[(3R,5S)-7-chloro-5-(2,3-dimethoxyphenyl)-1-(2,2-dimethylpropyl)-2-oxo-1,2,3,5-tetrahydro-4,1-benzoxazepin-3-yl]acetyl}-L-aspartic acid, PHOSPHATE ION, Squalene synthase | | Authors: | Suzuki, M, Shimizu, H, Katakura, S, Yamazaki, K, Higashihashi, N, Ichikawa, M, Yokomizo, A, Itoh, M, Sugita, K, Usui, H. | | Deposit date: | 2010-12-21 | | Release date: | 2011-12-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a new 2-aminobenzhydrol template for highly potent squalene synthase inhibitors
Bioorg.Med.Chem., 19, 2011
|
|
3Q30
 
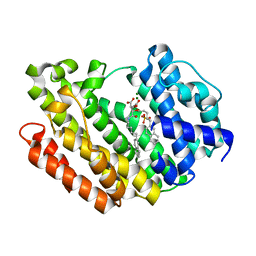 | | Human Squalene synthase in complex with (2R,3R)-2-Carboxymethoxy-3-[5-(2-naphthalenyl)pentyl]aminocarbonyl-3-[5-(2-naphthalenyl)pentyloxy]propionic acid | | Descriptor: | (2R,3R)-2-(carboxymethoxy)-4-{[5-(naphthalen-2-yl)pentyl]amino}-3-{[5-(naphthalen-2-yl)pentyl]oxy}-4-oxobutanoic acid, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Suzuki, M, Shimizu, H, Katakura, S, Yamazaki, K, Higashihashi, N, Ichikawa, M, Yokomizo, A, Itoh, M, Sugita, K, Usui, H. | | Deposit date: | 2010-12-21 | | Release date: | 2011-12-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a new 2-aminobenzhydrol template for highly potent squalene synthase inhibitors
Bioorg.Med.Chem., 19, 2011
|
|
1XSS
 
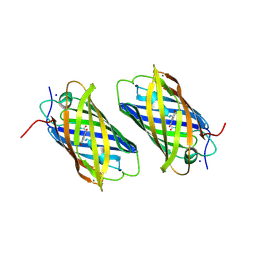 | | Semi-rational engineering of a green-emitting coral fluorescent protein into an efficient highlighter. | | Descriptor: | MAGNESIUM ION, SODIUM ION, fluorescent protein | | Authors: | Tsutsui, H, Karasawa, S, Shimizu, H, Nukina, N, Miyawaki, A. | | Deposit date: | 2004-10-20 | | Release date: | 2005-03-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Semi-rational engineering of a coral fluorescent protein into an efficient highlighter
Embo Rep., 6, 2005
|
|
1ZBZ
 
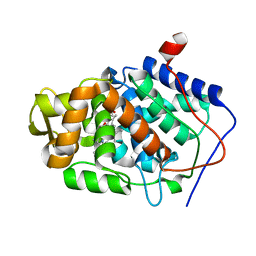 | | High-Resolution Crystal Structure of Compound I intermediate of Cytochrome c Peroxidase (CcP) | | Descriptor: | Cytochrome c peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bonagura, C.A, Bhaskar, B, Shimizu, H, Li, H, Sundaramoorthy, M, McRee, D.E, Goodin, D.B, Poulos, T.L. | | Deposit date: | 2005-04-09 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | High-resolution crystal structures and spectroscopy of native and compound I cytochrome c peroxidase
Biochemistry, 42, 2003
|
|
1ZBY
 
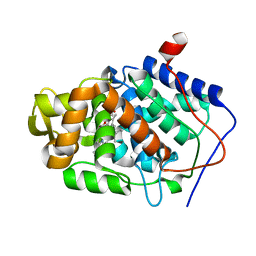 | | High-Resolution Crystal Structure of Native (Resting) Cytochrome c Peroxidase (CcP) | | Descriptor: | Cytochrome c peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bonagura, C.A, Bhaskar, B, Shimizu, H, Li, H, Sundaramoorthy, M, McRee, D.E, Goodin, D.B, Poulos, T.L. | | Deposit date: | 2005-04-09 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution crystal structures and spectroscopy of native and compound I cytochrome c peroxidase
Biochemistry, 42, 2003
|
|
1PKF
 
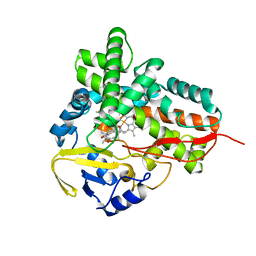 | | Crystal Structure of Epothilone D-bound Cytochrome P450epoK | | Descriptor: | EPOTHILONE D, PROTOPORPHYRIN IX CONTAINING FE, cytochrome p450EpoK | | Authors: | Nagano, S, Li, H, Shimizu, H, Nishida, C, Ogura, H, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2003-06-05 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Epothilone D-bound, Epothilone B-bound, and Substrate-free Forms of Cytochrome P450epoK
J.Biol.Chem., 278, 2003
|
|
1Q5E
 
 | | Substrate-free Cytochrome P450epoK | | Descriptor: | P450 epoxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagano, S, Li, H, Shimizu, H, Nishida, C, Ogura, H, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2003-08-06 | | Release date: | 2003-10-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structures of epothilone D-bound, epothilone B-bound, and substrate-free forms of cytochrome P450epoK
J.Biol.Chem., 278, 2003
|
|
1Q5D
 
 | | Epothilone B-bound Cytochrome P450epoK | | Descriptor: | 7,11-DIHYDROXY-8,8,10,12,16-PENTAMETHYL-3-[1-METHYL-2-(2-METHYL-THIAZOL-4-YL)VINYL]-4,17-DIOXABICYCLO[14.1.0]HEPTADECANE-5,9-DIONE, P450 epoxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagano, S, Li, H, Shimizu, H, Nishida, C, Ogura, H, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2003-08-06 | | Release date: | 2003-10-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of epothilone D-bound, epothilone B-bound, and substrate-free forms of cytochrome P450epoK
J.Biol.Chem., 278, 2003
|
|
1IFA
 
 | | THREE-DIMENSIONAL CRYSTAL STRUCTURE OF RECOMBINANT MURINE INTERFERON-BETA | | Descriptor: | ASPARAGINE, INTERFERON-BETA | | Authors: | Mitsui, Y, Senda, T, Matsuda, S, Kawano, G, Nakamura, K.T, Shimizu, H. | | Deposit date: | 1991-10-29 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-dimensional crystal structure of recombinant murine interferon-beta.
EMBO J., 11, 1992
|
|
1OM4
 
 | | STRUCTURE OF RAT NEURONAL NOS HEME DOMAIN WITH L-ARGININE BOUND | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ARGININE, ... | | Authors: | Li, H, Martasek, P, Shimizu, H, Masters, B.S.S, Poulos, T.L, Raman, C.S. | | Deposit date: | 2003-02-24 | | Release date: | 2003-03-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of Rat Neuronal NOS Heme Domain with L-Arginine Bound
To be Published
|
|
7VL2
 
 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with ethyl alpha-D-Glucoside | | Descriptor: | (2~{S},3~{R},4~{S},5~{S},6~{R})-2-ethoxy-6-(hydroxymethyl)oxane-3,4,5-triol, CALCIUM ION, beta-1,2-glucosyltransferase | | Authors: | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | Deposit date: | 2021-10-01 | | Release date: | 2022-03-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J.Biol.Chem., 298, 2022
|
|
7VKX
 
 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with glucose | | Descriptor: | CALCIUM ION, beta-1,2-glucosyltransferase, beta-D-glucopyranose | | Authors: | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | Deposit date: | 2021-10-01 | | Release date: | 2022-03-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J.Biol.Chem., 298, 2022
|
|
7VL5
 
 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with n-octyl-beta-D-glucoside | | Descriptor: | Beta-galactosidase, CALCIUM ION, octyl beta-D-glucopyranoside | | Authors: | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | Deposit date: | 2021-10-01 | | Release date: | 2022-03-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J.Biol.Chem., 298, 2022
|
|
7VKW
 
 | | The apo structure of beta-1,2-glucosyltransferase from Ignavibacterium album | | Descriptor: | beta-1,2-glucosyltransferase | | Authors: | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | Deposit date: | 2021-10-01 | | Release date: | 2022-03-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J.Biol.Chem., 298, 2022
|
|
7VL7
 
 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with esculin | | Descriptor: | 6-[(2S,3R,4S,5S,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-7-oxidanyl-chromen-2-one, CALCIUM ION, beta-1,2-glucosyltransferase | | Authors: | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | Deposit date: | 2021-10-01 | | Release date: | 2022-03-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J.Biol.Chem., 298, 2022
|
|
