1MBG
 
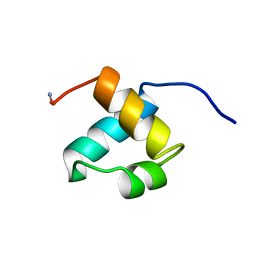 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 2 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBK
 
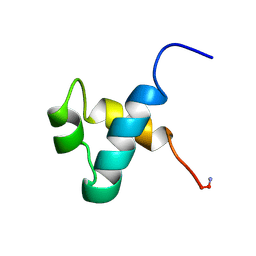 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 3 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
4WY3
 
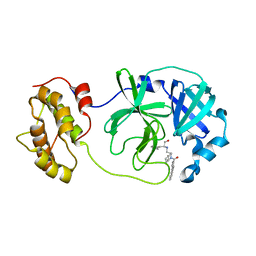 | | Structure of SARS-3CL protease complex with a phenylbenzoyl (R,S)-N-decalin type inhibitor | | Descriptor: | (2S)-2-({[(3R,4aS,8aR)-2-(biphenyl-4-ylcarbonyl)decahydroisoquinolin-3-yl]methyl}amino)-3-(1H-imidazol-5-yl)propanal, 3C-like proteinase | | Authors: | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2014-11-15 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors.
Bioorg.Med.Chem., 23, 2015
|
|
8I53
 
 | |
7W9Q
 
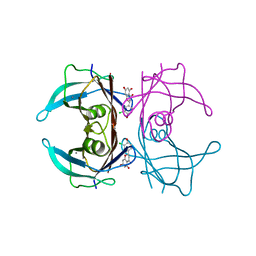 | | Crystal structure of V30M-TTR in complex with naringenin derivative-14 | | Descriptor: | (2~{R})-2-(3-chloranyl-4-oxidanyl-phenyl)-5,7-bis(oxidanyl)-2,3-dihydrochromen-4-one, CALCIUM ION, Transthyretin | | Authors: | Katayama, W, Shimane, A, Nabeshima, Y, Yokoyama, T, Mizuguchi, M. | | Deposit date: | 2021-12-10 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Chlorinated Naringenin Analogues as Potential Inhibitors of Transthyretin Amyloidogenesis.
J.Med.Chem., 65, 2022
|
|
7W9R
 
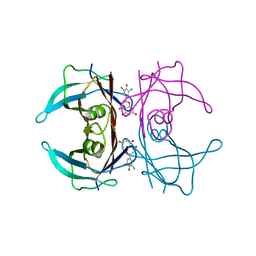 | | Crystal structure of V30M-TTR in complex with naringenin derivative-18 | | Descriptor: | (2~{R})-2-[3,5-bis(chloranyl)-4-oxidanyl-phenyl]-5,7-bis(oxidanyl)-2,3-dihydrochromen-4-one, Transthyretin | | Authors: | Katayama, W, Shimane, A, Nabeshima, Y, Yokoyama, T, Mizuguchi, M. | | Deposit date: | 2021-12-10 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Chlorinated Naringenin Analogues as Potential Inhibitors of Transthyretin Amyloidogenesis.
J.Med.Chem., 65, 2022
|
|
1MBF
 
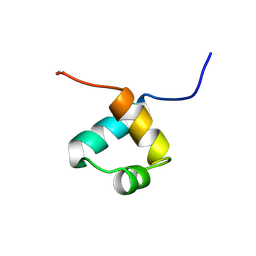 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 1 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
2CZY
 
 | | Solution structure of the NRSF/REST-mSin3B PAH1 complex | | Descriptor: | Paired amphipathic helix protein Sin3b, transcription factor REST (version 3) | | Authors: | Nomura, M, Uda-Tochio, H, Murai, K, Mori, N, Nishimura, Y. | | Deposit date: | 2005-07-20 | | Release date: | 2005-12-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Neural Repressor NRSF/REST Binds the PAH1 Domain of the Sin3 Corepressor by Using its Distinct Short Hydrophobic Helix
J.Mol.Biol., 354, 2005
|
|
5H0Q
 
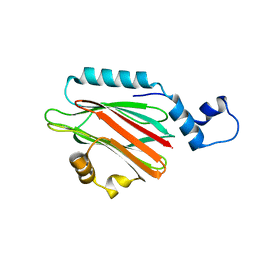 | | Crystal structure of lipid binding protein Nakanori at 1.5A | | Descriptor: | Lipid binding protein | | Authors: | Makino, A, Abe, M, Ishitsuka, R, Murate, M, Kishimoto, T, Sakai, S, Hullin-Matsuda, F, Shimada, Y, Inaba, T, Miyatake, H, Tanaka, H, Kurahashi, A, Pack, C.G, Kasai, R.S, Kubo, S, Schieber, N.L, Dohmae, N, Tochio, N, Hagiwara, K, Sasaki, Y, Aida, Y, Fujimori, F, Kigawa, T, Nishikori, K, Parton, R.G, Kusumi, A, Sako, Y, Anderluh, G, Yamashita, M, Kobayashi, T, Greimel, P, Kobayashi, T. | | Deposit date: | 2016-10-06 | | Release date: | 2016-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | A novel sphingomyelin/cholesterol domain-specific probe reveals the dynamics of the membrane domains during virus release and in Niemann-Pick type C
FASEB J., 31, 2017
|
|
1EWV
 
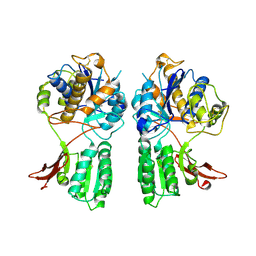 | | CRYSTAL STRUCTURE OF METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 LIGAND FREE FORM II | | Descriptor: | METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 | | Authors: | Kunishima, N, Shimada, Y, Tsuji, Y, Jingami, H, Morikawa, K. | | Deposit date: | 2000-04-27 | | Release date: | 2000-12-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor.
Nature, 407, 2000
|
|
5GOW
 
 | |
3VOV
 
 | | Crystal Structure of ROK Hexokinase from Thermus thermophilus | | Descriptor: | GLYCEROL, Glucokinase, ZINC ION | | Authors: | Nakamura, T, Kashima, Y, Mine, S, Oku, T, Uegaki, K. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Characterization and crystal structure of the thermophilic ROK hexokinase from Thermus thermophilus
J.Biosci.Bioeng., 2012
|
|
2DPT
 
 | |
2DPS
 
 | |
2E8J
 
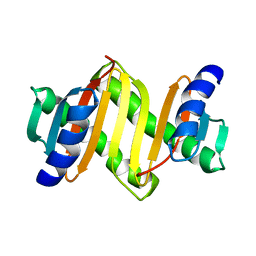 | |
3WJ2
 
 | | Crystal structure of ESTFA (FE-lacking apo form) | | Descriptor: | Carboxylesterase | | Authors: | Ohara, K, Unno, H, Oshima, Y, Furukawa, K, Fujino, N, Hirooka, K, Hemmi, H, Takahashi, S, Nishino, T, Kusunoki, M, Nakayama, T. | | Deposit date: | 2013-10-03 | | Release date: | 2014-07-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural insights into the low pH adaptation of a unique carboxylesterase from Ferroplasma: altering the pH optima of two carboxylesterases.
J.Biol.Chem., 289, 2014
|
|
1EWT
 
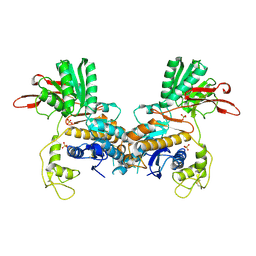 | | CRYSTAL STRUCTURE OF METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 LIGAND FREE FORM I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1, SULFATE ION | | Authors: | Kunishima, N, Shimada, Y, Tsuji, Y, Jingami, H, Morikawa, K. | | Deposit date: | 2000-04-27 | | Release date: | 2000-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor.
Nature, 407, 2000
|
|
2DY7
 
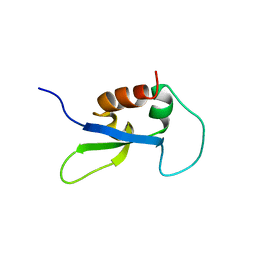 | |
2Z33
 
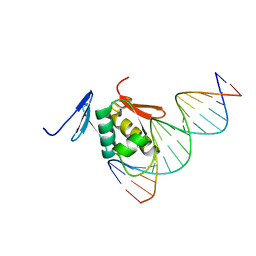 | | Solution structure of the DNA complex of PhoB DNA-binding/transactivation Domain | | Descriptor: | 5'-D(*AP*CP*AP*GP*AP*TP*TP*TP*AP*TP*GP*AP*CP*AP*GP*T)-3', 5'-D(*AP*CP*TP*GP*TP*CP*AP*TP*AP*AP*AP*TP*CP*TP*GP*T)-3', Phosphate regulon transcriptional regulatory protein phoB | | Authors: | Yamane, T, Okamura, H, Ikeguchi, M, Nishimura, Y, Kidera, A. | | Deposit date: | 2007-05-31 | | Release date: | 2008-04-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Water-mediated interactions between DNA and PhoB DNA-binding/transactivation domain: NMR-restrained molecular dynamics in explicit water environment.
Proteins, 71, 2008
|
|
3WD7
 
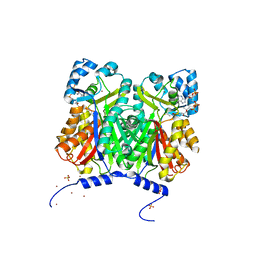 | | Type III polyketide synthase | | Descriptor: | COENZYME A, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Mori, T, Shimokawa, Y, Matsui, T, Kato, R, Sugio, S, Morita, H, Abe, I. | | Deposit date: | 2013-06-10 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Cloning, characterization, and crystal structure analysis of novel type III polyketide synthases from Citrus microcarpa
To be Published
|
|
3A5P
 
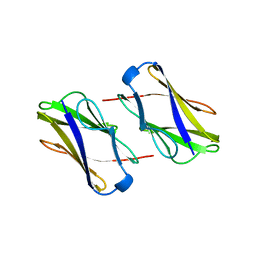 | | Crystal structure of hemagglutinin | | Descriptor: | Haemagglutinin I | | Authors: | Watanabe, N, Sakai, N, Nakamura, T, Nabeshima, Y, Kouno, T, Mizuguchi, M, Kawano, K. | | Deposit date: | 2009-08-10 | | Release date: | 2010-08-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Structure of Physarum polycephalum hemagglutinin I suggests a minimal carbohydrate recognition domain of legume lectin fold
J.Mol.Biol., 405, 2011
|
|
3WD8
 
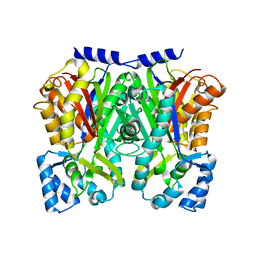 | | TypeIII polyketide synthases | | Descriptor: | GLYCEROL, Type III polyketide synthase quinolone synthase | | Authors: | Mori, T, Shimokawa, Y, Matsui, T, Morita, H, Abe, I. | | Deposit date: | 2013-06-10 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.463 Å) | | Cite: | Cloning, characterization, and crystal structure analysis of novel type III polyketide synthases from Citrus microcarpa
To be Published
|
|
2DY8
 
 | |
5C5O
 
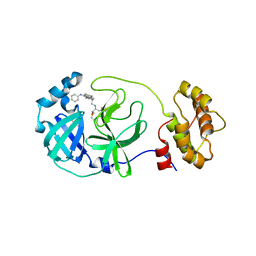 | | Structure of SARS-3CL protease complex with a phenyl-beta-alanyl (S,R)-N-decalin type inhibitor | | Descriptor: | (2S)-3-(1H-imidazol-5-yl)-2-({[(3S,4aR,8aS)-2-(N-phenyl-beta-alanyl)decahydroisoquinolin-3-yl]methyl}amino)propanal, 3C-like proteinase | | Authors: | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2015-06-21 | | Release date: | 2016-06-22 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Fused-ring structure of N-decalin as a novel scaffold for SARS 3CL protease inhibitors
to be published
|
|
8I75
 
 | |
