5ZFU
 
 | | Structure of the ExbB/ExbD hexameric complex (ExbB6ExbD3TM) | | Descriptor: | 22-mer peptide from Biopolymer transport protein ExbD, Biopolymer transport protein ExbB | | Authors: | Yonekura, K, Yamashita, Y, Matsuoka, R, Maki-Yonekura, S. | | Deposit date: | 2018-03-07 | | Release date: | 2018-05-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Hexameric and pentameric complexes of the ExbBD energizer in the Ton system.
Elife, 7, 2018
|
|
5ZFV
 
 | | Structure of the ExbB/ExbD pentameric complex (ExbB5ExbD1TM) | | Descriptor: | 22-mer peptide from Biopolymer transport protein ExbD, Biopolymer transport protein ExbB | | Authors: | Yonekura, K, Yamashita, Y, Matsuoka, R, Maki-Yonekura, S. | | Deposit date: | 2018-03-07 | | Release date: | 2018-05-09 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Hexameric and pentameric complexes of the ExbBD energizer in the Ton system.
Elife, 7, 2018
|
|
3AGT
 
 | | Hemerythrin-like domain of DcrH (met) | | Descriptor: | CHLORO DIIRON-OXO MOIETY, Hemerythrin-like domain protein DcrH | | Authors: | Onoda, A, Okamoto, Y, Sugimoto, H, Mizohata, E, Inoue, T, Shiro, Y, Hayashi, T. | | Deposit date: | 2010-04-06 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characteristics of Diiron Site with Large Cavity in Hemerythrin-like Domain of DcrH
to be published
|
|
1UC4
 
 | | Structure of diol dehydratase complexed with (S)-1,2-propanediol | | Descriptor: | AMMONIUM ION, CYANOCOBALAMIN, POTASSIUM ION, ... | | Authors: | Shibata, N, Nakanishi, Y, Fukuoka, M, Yamanishi, M, Yasuoka, N, Toraya, T. | | Deposit date: | 2003-04-08 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural rationalization for the lack of stereospecificity in coenzyme B12-dependent diol dehydratase
J.BIOL.CHEM., 278, 2003
|
|
1UC5
 
 | | Structure of diol dehydratase complexed with (R)-1,2-propanediol | | Descriptor: | AMMONIUM ION, CYANOCOBALAMIN, POTASSIUM ION, ... | | Authors: | Shibata, N, Nakanishi, Y, Fukuoka, M, Yamanishi, M, Yasuoka, N, Toraya, T. | | Deposit date: | 2003-04-08 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural rationalization for the lack of stereospecificity in coenzyme B12-dependent diol dehydratase
J.Biol.Chem., 278, 2003
|
|
5XNZ
 
 | | Crystal structure of CreD complex with fumarate | | Descriptor: | CreD, FUMARIC ACID | | Authors: | Katsuyama, Y, Sato, Y, Sugai, Y, Higashiyama, Y, Senda, M, Senda, T, Ohnishi, Y. | | Deposit date: | 2017-05-25 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the nitrosuccinate lyase CreD in complex with fumarate provides insights into the catalytic mechanism for nitrous acid elimination
FEBS J., 285, 2018
|
|
5Z76
 
 | | Artificial L-threonine 3-dehydrogenase designed by full consensus design | | Descriptor: | Artificial L-threonine 3-dehydrogenase | | Authors: | Nakano, S, Motoyama, T, Miyashita, Y, Ishizuka, Y, Matsuo, N, Tokiwa, H, Shinoda, S, Asano, Y, Ito, S. | | Deposit date: | 2018-01-27 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Benchmark Analysis of Native and Artificial NAD+-Dependent Enzymes Generated by a Sequence-Based Design Method with or without Phylogenetic Data.
Biochemistry, 57, 2018
|
|
3ALJ
 
 | | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, reduced form | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kobayashi, J, Yoshida, H, Yoshikane, Y, Kamitori, S, Yagi, T. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
5XNY
 
 | | Crystal structure of CreD | | Descriptor: | CreD | | Authors: | Katsuyama, Y, Sato, Y, Sugai, Y, Higashiyama, Y, Senda, M, Senda, T, Ohnishi, Y. | | Deposit date: | 2017-05-25 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal structure of the nitrosuccinate lyase CreD in complex with fumarate provides insights into the catalytic mechanism for nitrous acid elimination
FEBS J., 285, 2018
|
|
5Z75
 
 | | Artificial L-threonine 3-dehydrogenase designed by ancestral sequence reconstruction. | | Descriptor: | Artificial L-threonine 3-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NONAETHYLENE GLYCOL, ... | | Authors: | Nakano, S, Motoyama, T, Miyashita, Y, Ishizuka, Y, Matsuo, N, Tokiwa, H, Shinoda, S, Asano, Y, Ito, S. | | Deposit date: | 2018-01-27 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Benchmark Analysis of Native and Artificial NAD+-Dependent Enzymes Generated by a Sequence-Based Design Method with or without Phylogenetic Data.
Biochemistry, 57, 2018
|
|
1V5H
 
 | | Crystal Structure of Human Cytoglobin (Ferric Form) | | Descriptor: | Cytoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugimoto, H, Makino, M, Sawai, H, Kawada, N, Yoshizato, K, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-23 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of human cytoglobin for ligand binding.
J.Mol.Biol., 339, 2004
|
|
3ALM
 
 | | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, mutant C294A | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Kobayashi, J, Yoshida, H, Yoshikane, Y, Kamitori, S, Yagi, T. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
5ZV2
 
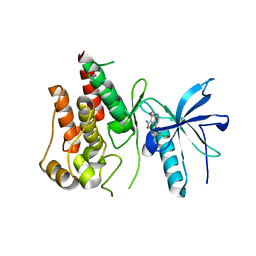 | | FGFR-1 in complex with ligand lenvatinib | | Descriptor: | 4-{3-chloro-4-[(cyclopropylcarbamoyl)amino]phenoxy}-7-methoxyquinoline-6-carboxamide, Fibroblast growth factor receptor 1 | | Authors: | Matsuki, M, Hoshi, T, Yamamoto, Y, Ikemori-Kawada, M, Minoshima, Y, Funahashi, Y, Matsui, J. | | Deposit date: | 2018-05-09 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Lenvatinib inhibits angiogenesis and tumor fibroblast growth factor signaling pathways in human hepatocellular carcinoma models.
Cancer Med, 7, 2018
|
|
6A8V
 
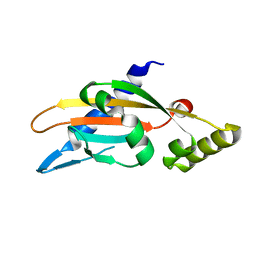 | | PhoQ sensor domain (D179R mutant): analysis of internal cavity | | Descriptor: | Sensor protein PhoQ | | Authors: | Yoshitani, K, Ishii, E, Taniguchi, K, Sugimoto, H, Shiro, Y, Mori, H, Akiyama, Y, Kato, A, Utsumi, R, Eguchi, Y. | | Deposit date: | 2018-07-10 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of an internal cavity in the PhoQ sensor domain for PhoQ activity and SafA-mediated control.
Biosci. Biotechnol. Biochem., 83, 2019
|
|
6A8U
 
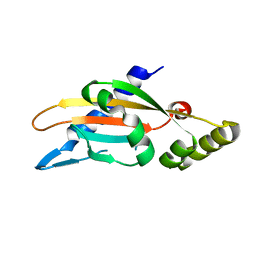 | | PhoQ sensor domain (wild type): analysis of internal cavity | | Descriptor: | Sensor protein PhoQ | | Authors: | Yoshitani, K, Ishii, E, Taniguchi, K, Sugimoto, H, Shiro, Y, Mori, H, Akiyama, Y, Kato, A, Utsumi, R, Eguchi, Y. | | Deposit date: | 2018-07-10 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Identification of an internal cavity in the PhoQ sensor domain for PhoQ activity and SafA-mediated control.
Biosci. Biotechnol. Biochem., 83, 2019
|
|
1VD4
 
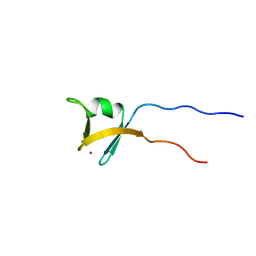 | | Solution structure of the zinc finger domain of TFIIE alpha | | Descriptor: | Transcription initiation factor IIE, alpha subunit, ZINC ION | | Authors: | Okuda, M, Tanaka, A, Arai, Y, Satoh, M, Okamura, H, Nagadoi, A, Hanaoka, F, Ohkuma, Y, Nishimura, Y. | | Deposit date: | 2004-03-18 | | Release date: | 2004-10-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel zinc finger structure in the large subunit of human general transcription factor TFIIE.
J.Biol.Chem., 279, 2004
|
|
3ALL
 
 | | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, mutant Y270A | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kobayashi, J, Yoshida, H, Yoshikane, Y, Kamitori, S, Yagi, T. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be published
|
|
2ZA6
 
 | | recombinant horse L-chain apoferritin | | Descriptor: | CADMIUM ION, Ferritin light chain | | Authors: | Yamashita, I, Mishima, Y, Park, S.-Y, Heddle, J.G, Tame, J.R.H. | | Deposit date: | 2007-10-02 | | Release date: | 2008-01-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Effect of N-terminal Residues on the Structural Stability of Recombinant Horse L-chain Apoferritin in an Acidic Environment
J.BIOCHEM.(TOKYO), 142, 2007
|
|
2ZA7
 
 | | recombinant horse L-chain apoferritin N-terminal deletion mutant (residues 1-4) | | Descriptor: | Ferritin light chain | | Authors: | Yamashita, I, Mishima, Y, Park, S.-Y, Heddle, J.G, Tame, J.R.H. | | Deposit date: | 2007-10-02 | | Release date: | 2008-01-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Effect of N-terminal Residues on the Structural Stability of Recombinant Horse L-chain Apoferritin in an Acidic Environment
J.BIOCHEM.(TOKYO), 142, 2007
|
|
3A0R
 
 | | Crystal structure of histidine kinase ThkA (TM1359) in complex with response regulator protein TrrA (TM1360) | | Descriptor: | MERCURY (II) ION, Response regulator, Sensor protein | | Authors: | Yamada, S, Sugimoto, H, Kobayashi, M, Ohno, A, Nakamura, H, Shiro, Y. | | Deposit date: | 2009-03-24 | | Release date: | 2009-10-20 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of PAS-linked histidine kinase and the response regulator complex
Structure, 17, 2009
|
|
5YQ0
 
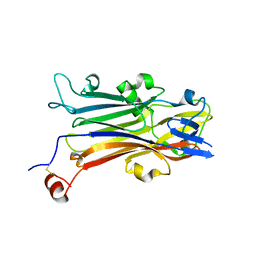 | | Crystal structure of secreted protein CofJ from ETEC. | | Descriptor: | CALCIUM ION, CofJ | | Authors: | Oki, H, Kawahara, K, Maruno, T, Imai, T, Muroga, Y, Fukakusa, S, Iwashita, T, Kobayashi, Y, Matsuda, S, Kodama, T, Iida, T, Yoshida, T, Ohkubo, T, Nakamura, S. | | Deposit date: | 2017-11-04 | | Release date: | 2018-06-27 | | Last modified: | 2018-07-25 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Interplay of a secreted protein with type IVb pilus for efficient enterotoxigenicEscherichia colicolonization
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YPZ
 
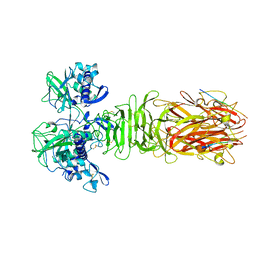 | | Crystal structure of minor pilin CofB from CFA/III complexed with N-terminal peptide fragment of CofJ | | Descriptor: | CofB, CofJ | | Authors: | Oki, H, Kawahara, K, Maruno, T, Imai, T, Muroga, Y, Fukakusa, S, Iwashita, T, Kobayashi, Y, Matsuda, S, Kodama, T, Iida, T, Yoshida, T, Ohkubo, T, Nakamura, S. | | Deposit date: | 2017-11-04 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.521 Å) | | Cite: | Interplay of a secreted protein with type IVb pilus for efficient enterotoxigenicEscherichia colicolonization.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2ZD0
 
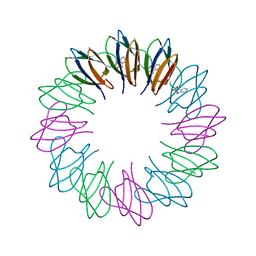 | | Crystal structures and thermostability of mutant TRAP3 A5 (ENGINEERED TRAP) | | Descriptor: | TRYPTOPHAN, Transcription attenuation protein mtrB | | Authors: | Watanabe, M, Mishima, Y, Yamashita, I, Park, S.Y, Tame, J.R.H, Heddle, J.G. | | Deposit date: | 2007-11-15 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Intersubunit linker length as a modifier of protein stability: crystal structures and thermostability of mutant TRAP.
Protein Sci., 17, 2008
|
|
2ZCZ
 
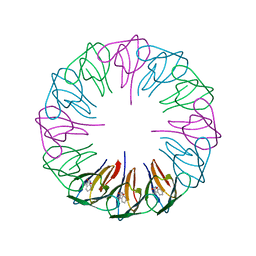 | | Crystal structures and thermostability of mutant TRAP3 A7 (ENGINEERED TRAP) | | Descriptor: | TRYPTOPHAN, Transcription attenuation protein mtrB | | Authors: | Watanabe, M, Mishima, Y, Yamashita, I, Park, S.Y, Tame, J.R.H, Heddle, J.G. | | Deposit date: | 2007-11-15 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Intersubunit linker length as a modifier of protein stability: crystal structures and thermostability of mutant TRAP.
Protein Sci., 17, 2008
|
|
5XHL
 
 | | Crystal Structure of HasAp with Gallium Phthalocyanine | | Descriptor: | Heme acquisition protein HasAp, Phthalocyanine containing GA | | Authors: | Shoji, O, Shisaka, Y, Iwai, Y, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2017-04-21 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of HasAp with Gallium Phthalocyanine
to be published
|
|
