6LK8
 
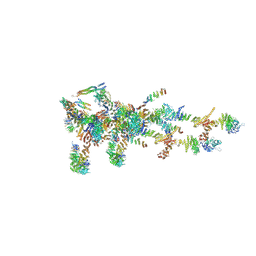 | | Structure of Xenopus laevis Cytoplasmic Ring subunit. | | Descriptor: | GATOR complex protein SEC13, MGC154553 protein, MGC83295 protein, ... | | Authors: | Shi, Y, Huang, G, Yan, C, Zhang, Y. | | Deposit date: | 2019-12-18 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex by cryo-electron microscopy single particle analysis.
Cell Res., 30, 2020
|
|
2YGS
 
 | | CARD DOMAIN FROM APAF-1 | | Descriptor: | APOPTOTIC PROTEASE ACTIVATING FACTOR 1 | | Authors: | Shi, Y. | | Deposit date: | 1999-05-08 | | Release date: | 2000-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of procaspase-9 recruitment by the apoptotic protease-activating factor 1.
Nature, 399, 1999
|
|
7CBT
 
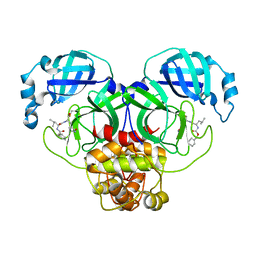 | | The crystal structure of SARS-CoV-2 main protease in complex with GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Shi, Y, Peng, G. | | Deposit date: | 2020-06-13 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.346 Å) | | Cite: | The preclinical inhibitor GS441524 in combination with GC376 efficaciously inhibited the proliferation of SARS-CoV-2 in the mouse respiratory tract.
Emerg Microbes Infect, 10, 2021
|
|
5YQ7
 
 | | Cryo-EM structure of the RC-LH core complex from Roseiflexus castenholzii | | Descriptor: | 2-methyl-3-[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,14,18,22,26,30,34,38,42-undecaen-1-yl]naphthalene-1,4-dione, Alpha subunit of light-harvesting 1, BACTERIOCHLOROPHYLL A, ... | | Authors: | Shi, Y, Xin, Y.Y, Niu, T.X, Wang, Q.Q, Niu, W.Q, Huang, X.J, Ding, W, Blankenship, R.E, Xu, X.L, Sun, F. | | Deposit date: | 2017-11-05 | | Release date: | 2018-05-02 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the RC-LH core complex from an early branching photosynthetic prokaryote.
Nat Commun, 9, 2018
|
|
1MHD
 
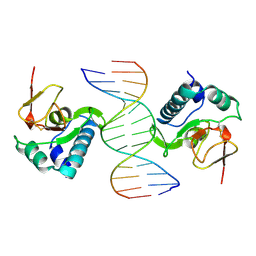 | | CRYSTAL STRUCTURE OF A SMAD MH1 DOMAIN BOUND TO DNA | | Descriptor: | DNA, SMAD3 | | Authors: | Shi, Y. | | Deposit date: | 1998-08-18 | | Release date: | 1999-08-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-beta signaling.
Cell(Cambridge,Mass.), 94, 1998
|
|
1YGS
 
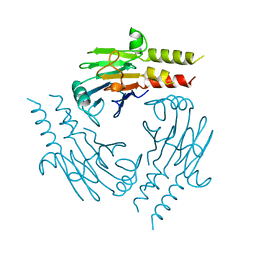 | | CRYSTAL STRUCTURE OF THE SMAD4 TUMOR SUPPRESSOR C-TERMINAL DOMAIN | | Descriptor: | SMAD4 | | Authors: | Shi, Y, Hata, A, Lo, R.S, Massague, J, Pavletich, N.P. | | Deposit date: | 1997-10-03 | | Release date: | 1998-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural basis for mutational inactivation of the tumour suppressor Smad4.
Nature, 388, 1997
|
|
7W7H
 
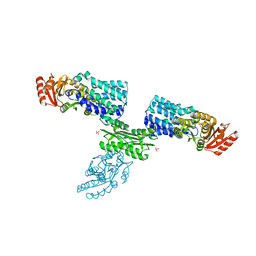 | | S Suis FakA-FakB2 complex structure | | Descriptor: | OLEIC ACID, Predicted kinase related to dihydroxyacetone kinase, SULFATE ION, ... | | Authors: | Shi, Y, Zang, N, Lou, N, Xu, Y, Sun, J, Huang, M, Zhang, H, Lu, H, Zhou, C, Feng, Y. | | Deposit date: | 2021-12-04 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and mechanism for streptococcal fatty acid kinase (Fak) system dedicated to host fatty acid scavenging.
Sci Adv, 8, 2022
|
|
7YER
 
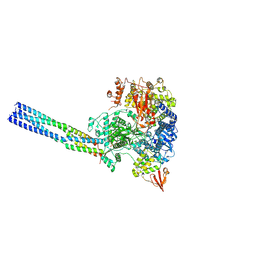 | | The structure of EBOV L-VP35 complex | | Descriptor: | Polymerase cofactor VP35, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Shi, Y, Yuan, B, Peng, Q. | | Deposit date: | 2022-07-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the Ebola virus polymerase complex.
Nature, 610, 2022
|
|
7YET
 
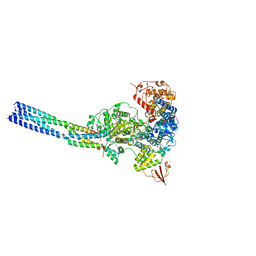 | | The structure of EBOV L-VP35 in complex with suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, Polymerase cofactor VP35, RNA-directed RNA polymerase L | | Authors: | Shi, Y, Yuan, B, Peng, Q. | | Deposit date: | 2022-07-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the Ebola virus polymerase complex.
Nature, 610, 2022
|
|
7YES
 
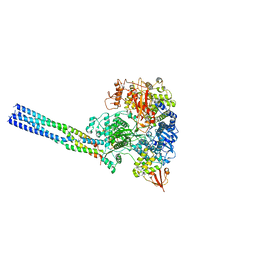 | |
5XOV
 
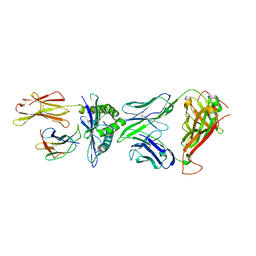 | | Crystal structure of peptide-HLA-A24 bound to S19-2 V-delta/V-beta TCR | | Descriptor: | Beta-2-microglobulin, HIV-1 Nef138-10 peptide, HLA class I histocompatibility antigen, ... | | Authors: | Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2017-05-31 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.684 Å) | | Cite: | Conserved V delta 1 Binding Geometry in a Setting of Locus-Disparate pHLA Recognition by delta / alpha beta T Cell Receptors (TCRs): Insight into Recognition of HIV Peptides by TCRs.
J. Virol., 91, 2017
|
|
5XOT
 
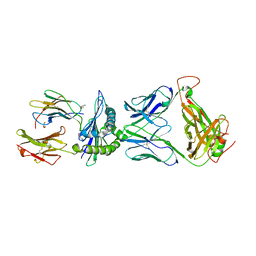 | | Crystal structure of pHLA-B35 in complex with TU55 T cell receptor | | Descriptor: | An HIV reverse transcriptase epitope, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2017-05-31 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.787 Å) | | Cite: | Conserved V delta 1 Binding Geometry in a Setting of Locus-Disparate pHLA Recognition by delta / alpha beta T Cell Receptors (TCRs): Insight into Recognition of HIV Peptides by TCRs.
J. Virol., 91, 2017
|
|
5XOS
 
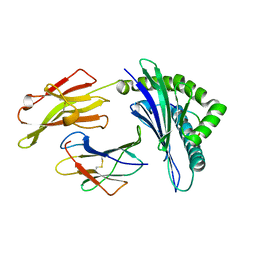 | | Crystal structure of HLA-B35 in complex with a pepetide antigen | | Descriptor: | An HIV reverse transcriptase epitope, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2017-05-31 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Conserved V delta 1 Binding Geometry in a Setting of Locus-Disparate pHLA Recognition by delta / alpha beta T Cell Receptors (TCRs): Insight into Recognition of HIV Peptides by TCRs.
J. Virol., 91, 2017
|
|
7FIK
 
 | | The cryo-EM structure of the CR subunit from X. laevis NPC | | Descriptor: | MGC154553 protein, MGC83295 protein, MGC83926 protein, ... | | Authors: | Shi, Y, Huang, G, Zhan, X. | | Deposit date: | 2021-07-31 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex.
Science, 376, 2022
|
|
7FIL
 
 | |
2RSE
 
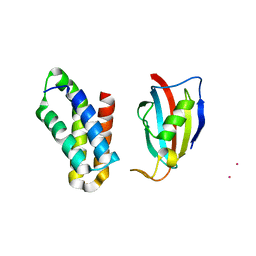 | | NMR structure of FKBP12-mTOR FRB domain-rapamycin complex structure determined based on PCS | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, Serine/threonine-protein kinase mTOR, TERBIUM(III) ION | | Authors: | Kobashigawa, Y, Ushio, M, Saio, T, Inagaki, F. | | Deposit date: | 2012-01-25 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Convenient method for resolving degeneracies due to symmetry of the magnetic susceptibility tensor and its application to pseudo contact shift-based protein-protein complex structure determination.
J.Biomol.Nmr, 53, 2012
|
|
1JDC
 
 | | MUTANT (E219Q) MALTOTETRAOSE-FORMING EXO-AMYLASE COCRYSTALLIZED WITH MALTOTETRAOSE (CRYSTAL TYPE 1) | | Descriptor: | 1,4-ALPHA MALTOTETRAHYDROLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yoshioka, Y, Hasegawa, K, Matsuura, Y, Katsube, Y, Kubota, M. | | Deposit date: | 1997-06-16 | | Release date: | 1997-10-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a mutant maltotetraose-forming exo-amylase cocrystallized with maltopentaose.
J.Mol.Biol., 271, 1997
|
|
1JDD
 
 | | MUTANT (E219Q) MALTOTETRAOSE-FORMING EXO-AMYLASE COCRYSTALLIZED WITH MALTOTETRAOSE (CRYSTAL TYPE 2) | | Descriptor: | 1,4-ALPHA MALTOTETRAHYDROLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yoshioka, Y, Hasegawa, K, Matsuura, Y, Katsube, Y, Kubota, M. | | Deposit date: | 1997-06-16 | | Release date: | 1997-10-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a mutant maltotetraose-forming exo-amylase cocrystallized with maltopentaose.
J.Mol.Biol., 271, 1997
|
|
1IZ0
 
 | |
1IYZ
 
 | |
5AVE
 
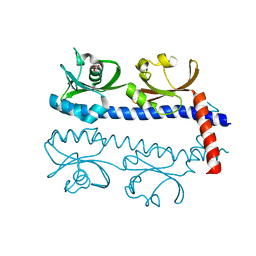 | | The ligand binding domain of Mlp37 with serine | | Descriptor: | Methyl-accepting chemotaxis (MCP) signaling domain protein, SERINE | | Authors: | Takahashi, Y, Sumita, K, Uchida, Y, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2015-06-15 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of a Vibrio cholerae chemoreceptor that senses taurine and amino acids as attractants
Sci Rep, 6, 2016
|
|
5AVF
 
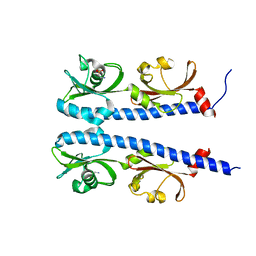 | | The ligand binding domain of Mlp37 with taurine | | Descriptor: | 2-AMINOETHANESULFONIC ACID, Methyl-accepting chemotaxis (MCP) signaling domain protein | | Authors: | Takahashi, Y, Sumita, K, Uchida, Y, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of a Vibrio cholerae chemoreceptor that senses taurine and amino acids as attractants
Sci Rep, 6, 2016
|
|
5ZQU
 
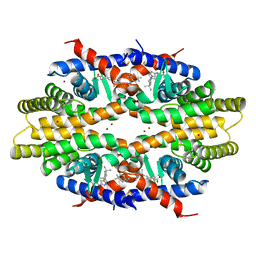 | | Crystal structure of tetrameric RXRalpha-LBD complexed with partial agonist CBt-PMN | | Descriptor: | 1-(3,5,5,8,8-pentamethyl-6,7-dihydronaphthalen-2-yl)benzotriazole-5-carboxylic acid, BROMIDE ION, Retinoic acid receptor RXR-alpha | | Authors: | Miyashita, Y, Numoto, N, Arulmozhiraja, S, Nakano, S, Matsuo, N, Shimizu, K, Kakuta, H, Ito, S, Ikura, T, Ito, N, Tokiwa, H. | | Deposit date: | 2018-04-20 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.60038781 Å) | | Cite: | Dual conformation of the ligand induces the partial agonistic activity of retinoid X receptor alpha (RXR alpha ).
FEBS Lett., 593, 2019
|
|
6AJN
 
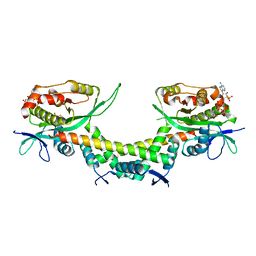 | | Crystal structure of AtaTR bound with AcCoA | | Descriptor: | ACETYL COENZYME *A, DUF1778 domain-containing protein, N-acetyltransferase | | Authors: | Yashiro, Y, Yamashita, S, Tomita, K. | | Deposit date: | 2018-08-28 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Crystal Structure of the Enterohemorrhagic Escherichia coli AtaT-AtaR Toxin-Antitoxin Complex.
Structure, 27, 2019
|
|
4XD7
 
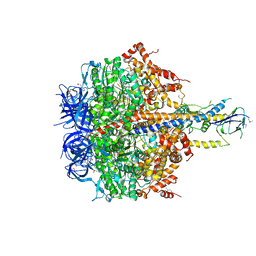 | | Structure of thermophilic F1-ATPase inhibited by epsilon subunit | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase epsilon chain, ATP synthase gamma chain, ... | | Authors: | SHIRAKIHARA, Y, SHIRATORI, A, TANIKAWA, H, NAKASAKO, M, YOSHIDA, M, SUZUKI, T. | | Deposit date: | 2014-12-19 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of a thermophilic F1 -ATPase inhibited by an epsilon-subunit: deeper insight into the epsilon-inhibition mechanism.
Febs J., 282, 2015
|
|
