6JUA
 
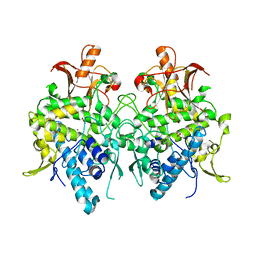 | | Aspergillus oryzae pro-tyrosinase oxygen-bound C92A mutant | | Descriptor: | COPPER (II) ION, PEROXIDE ION, Tyrosinase | | Authors: | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-04-13 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6JU4
 
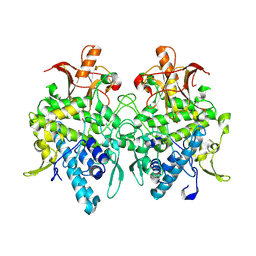 | | Aspergillus oryzae pro-tyrosinase F513Y mutant | | Descriptor: | COPPER (II) ION, Tyrosinase | | Authors: | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-04-13 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6JU8
 
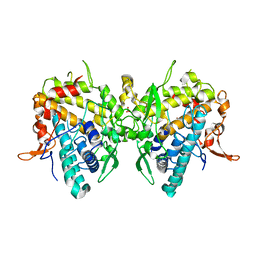 | | Aspergillus oryzae active-tyrosinase copper-bound C92A mutant | | Descriptor: | COPPER (II) ION, NITRATE ION, Tyrosinase | | Authors: | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-04-13 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6JUB
 
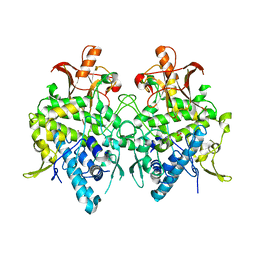 | | Radiation damage in Aspergillus oryzae pro-tyrosinase oxygen-bound C92A mutant | | Descriptor: | COPPER (II) ION, PEROXIDE ION, Tyrosinase | | Authors: | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-04-13 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
4UX1
 
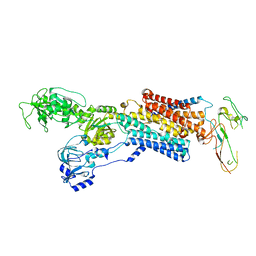 | | Cryo-EM structure of antagonist-bound E2P gastric H,K-ATPase (SCH.E2. AlF) | | Descriptor: | POTASSIUM-TRANSPORTING ATPASE ALPHA CHAIN 1, POTASSIUM-TRANSPORTING ATPASE SUBUNIT BETA | | Authors: | Abe, K, Tani, K, Fujiyoshi, Y. | | Deposit date: | 2014-08-18 | | Release date: | 2014-09-17 | | Last modified: | 2014-11-12 | | Method: | ELECTRON CRYSTALLOGRAPHY (8 Å) | | Cite: | Systematic Comparison of Molecular Conformations of H+,K+-ATPase Reveals an Important Contribution of the A-M2 Linker for the Luminal Gating.
J.Biol.Chem., 289, 2014
|
|
1AYG
 
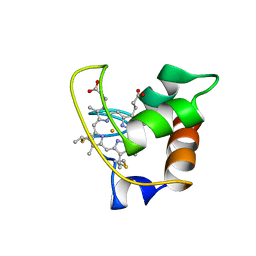 | | SOLUTION STRUCTURE OF CYTOCHROME C-552, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C-552, HEME C | | Authors: | Hasegawa, J, Yoshida, T, Yamazaki, T, Sambongi, Y, Yu, Y, Igarashi, Y, Kodama, T, Yamazaki, K, Hakusui, H, Kyogoku, Y, Kobayashi, Y. | | Deposit date: | 1997-11-04 | | Release date: | 1998-11-25 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of thermostable cytochrome c-552 from Hydrogenobacter thermophilus determined by 1H-NMR spectroscopy.
Biochemistry, 37, 1998
|
|
4P79
 
 | | Crystal structure of mouse claudin-15 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Claudin-15 | | Authors: | Suzuki, H, Nishizawa, T, Tani, K, Yamazaki, Y, Tamura, A, Ishitani, R, Dohmae, N, Tsukita, S, Nureki, O, Fujiyoshi, Y. | | Deposit date: | 2014-03-26 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a claudin provides insight into the architecture of tight junctions.
Science, 344, 2014
|
|
1IS1
 
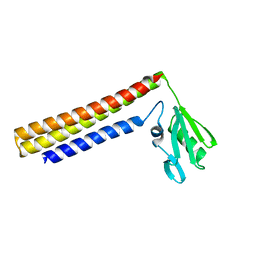 | | Crystal structure of ribosome recycling factor from Vibrio parahaemolyticus | | Descriptor: | RIBOSOME RECYCLING FACTOR | | Authors: | Nakano, H, Yamaichi, Y, Uchiyama, S, Yoshida, T, Nishina, K, Kato, H, Ohkubo, T, Honda, T, Yamagata, Y, Kobayashi, Y. | | Deposit date: | 2001-11-05 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and binding mode of a ribosome recycling factor (RRF) from mesophilic bacterium
J.BIOL.CHEM., 278, 2003
|
|
1D8K
 
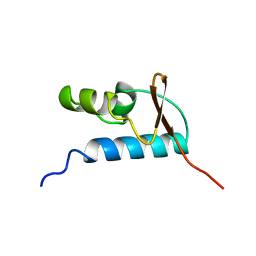 | | SOLUTION STRUCTURE OF THE CENTRAL CORE DOMAIN OF TFIIE BETA | | Descriptor: | GENERAL TRANSCRIPTION FACTOR TFIIE-BETA | | Authors: | Okuda, M, Watanabe, Y, Okamura, H, Hanaoka, F, Ohkuma, Y, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-25 | | Release date: | 2000-04-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the central core domain of TFIIEbeta with a novel double-stranded DNA-binding surface.
EMBO J., 19, 2000
|
|
2RNR
 
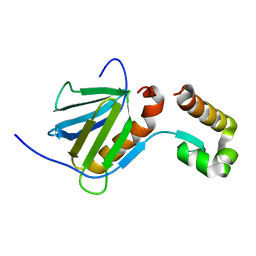 | |
1JF6
 
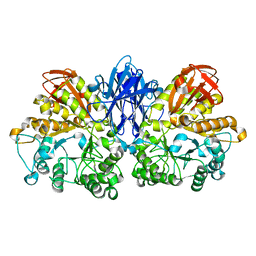 | | Crystal structure of thermoactinomyces vulgaris r-47 alpha-amylase mutant F286Y | | Descriptor: | ALPHA AMYLASE II, CALCIUM ION | | Authors: | Ohtaki, A, Kondo, S, Shimura, Y, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2001-06-20 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Role of Phe286 in the recognition mechanism of cyclomaltooligosaccharides (cyclodextrins) by Thermoactinomyces vulgaris R-47 alpha-amylase 2 (TVAII). X-ray structures of the mutant TVAIIs, F286A and F286Y, and kinetic analyses of the Phe286-replaced mutant TVAIIs
CARBOHYDR.RES., 334, 2001
|
|
1D8J
 
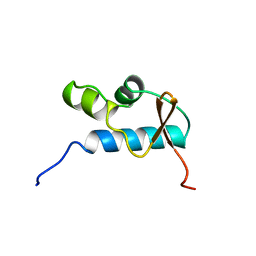 | | SOLUTION STRUCTURE OF THE CENTRAL CORE DOMAIN OF TFIIE BETA | | Descriptor: | GENERAL TRANSCRIPTION FACTOR TFIIE-BETA | | Authors: | Okuda, M, Watanabe, Y, Okamura, H, Hanaoka, F, Ohkuma, Y, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-25 | | Release date: | 2000-04-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the central core domain of TFIIEbeta with a novel double-stranded DNA-binding surface.
EMBO J., 19, 2000
|
|
5AZ2
 
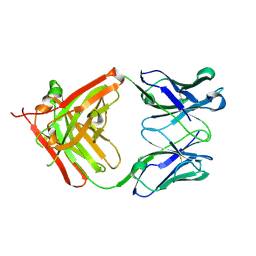 | | Crystal structure of the Fab fragment of 9E5, a murine monoclonal antibody specific for human epiregulin | | Descriptor: | anti-human epiregulin antibody 9E5 Fab heavy chain, anti-human epiregulin antibody 9E5 Fab light chain | | Authors: | Kado, Y, Mizohata, E, Nagatoishi, S, Iijima, M, Shinoda, K, Miyafusa, T, Nakayama, T, Yoshizumi, T, Sugiyama, A, Kawamura, T, Lee, Y.H, Matsumura, H, Doi, H, Fujitani, H, Kodama, T, Shibasaki, Y, Tsumoto, K, Inoue, T. | | Deposit date: | 2015-09-16 | | Release date: | 2015-12-09 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Epiregulin Recognition Mechanisms by Anti-epiregulin Antibody 9E5: STRUCTURAL, FUNCTIONAL, AND MOLECULAR DYNAMICS SIMULATION ANALYSES
J.Biol.Chem., 291, 2016
|
|
5X93
 
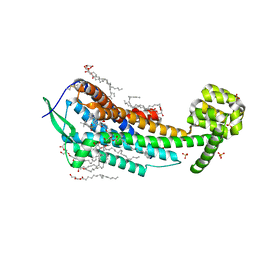 | | Human endothelin receptor type-B in complex with antagonist K-8794 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[6-[(4-tert-butylphenyl)sulfonylamino]-5-(2-methoxyphenoxy)-2-pyrimidin-2-yl-pyrimidin-4-yl]oxy-N-(2,6-dimethylphenyl)propanamide, CHOLESTEROL, ... | | Authors: | Shihoya, W, Nishizawa, T, Yamashita, K, Hirata, K, Okuta, A, Tani, K, Fujiyoshi, Y, Doi, T, Nureki, O. | | Deposit date: | 2017-03-05 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structures of endothelin ETB receptor bound to clinical antagonist bosentan and its analog
Nat. Struct. Mol. Biol., 24, 2017
|
|
1A0A
 
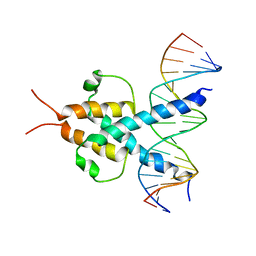 | | PHOSPHATE SYSTEM POSITIVE REGULATORY PROTEIN PHO4/DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*TP*AP*GP*TP*CP*CP*CP*AP*CP*GP*TP*GP*TP*GP*AP*G )-3'), DNA (5'-D(*CP*TP*CP*AP*CP*AP*CP*GP*TP*GP*GP*GP*AP*CP*TP*AP*G )-3'), PROTEIN (PHOSPHATE SYSTEM POSITIVE REGULATORY PROTEIN PHO4) | | Authors: | Shimizu, T, Toumoto, A, Ihara, K, Shimizu, M, Kyogoku, Y, Ogawa, N, Oshima, Y, Hakoshima, T. | | Deposit date: | 1997-11-27 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition.
EMBO J., 16, 1997
|
|
1GEB
 
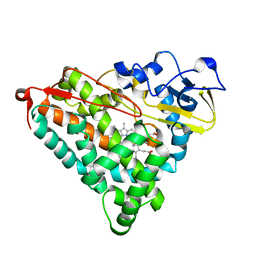 | | X-RAY CRYSTAL STRUCTURE AND CATALYTIC PROPERTIES OF THR252ILE MUTANT OF CYTOCHROME P450CAM | | Descriptor: | CAMPHOR, CYTOCHROME P450-CAM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hishiki, T, Shimada, H, Nagano, S, Park, S.-Y, Ishimura, Y. | | Deposit date: | 2000-11-01 | | Release date: | 2000-11-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | X-ray crystal structure and catalytic properties of Thr252Ile mutant of cytochrome P450cam: roles of Thr252 and water in the active center.
J.Biochem., 128, 2000
|
|
5XPR
 
 | | Human endothelin receptor type-B in complex with antagonist bosentan | | Descriptor: | 4-tert-butyl-N-[6-(2-hydroxyethyloxy)-5-(2-methoxyphenoxy)-2-pyrimidin-2-yl-pyrimidin-4-yl]benzenesulfonamide, Endothelin B receptor,Endolysin,Endothelin B receptor, SULFATE ION | | Authors: | Shihoya, W, Nishizawa, T, Yamashita, K, Hirata, K, Okuta, A, Tani, K, Fujiyoshi, Y, Doi, T, Nureki, O. | | Deposit date: | 2017-06-04 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | X-ray structures of endothelin ETB receptor bound to clinical antagonist bosentan and its analog
Nat. Struct. Mol. Biol., 24, 2017
|
|
1IYL
 
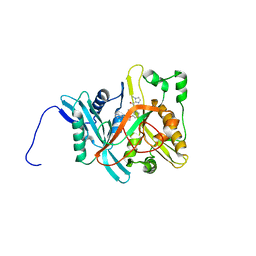 | | Crystal Structure of Candida albicans N-myristoyltransferase with Non-peptidic Inhibitor | | Descriptor: | (1-METHYL-1H-IMIDAZOL-2-YL)-(3-METHYL-4-{3-[(PYRIDIN-3-YLMETHYL)-AMINO]-PROPOXY}-BENZOFURAN-2-YL)-METHANONE, Myristoyl-CoA:Protein N-Myristoyltransferase | | Authors: | Sogabe, S, Fukami, T.A, Morikami, K, Shiratori, Y, Aoki, Y, D'Arcy, A, Winkler, F.K, Banner, D.W, Ohtsuka, T. | | Deposit date: | 2002-08-29 | | Release date: | 2002-12-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structures of Candida albicans N-Myristoyltransferase with Two Distinct Inhibitors
CHEM.BIOL., 9, 2002
|
|
1J1G
 
 | | Crystal structure of the RNase MC1 mutant N71S in complex with 5'-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Ribonuclease MC1 | | Authors: | Numata, T, Suzuki, A, Kakuta, Y, Kimura, K, Yao, M, Tanaka, I, Yoshida, Y, Ueda, T, Kimura, M. | | Deposit date: | 2002-12-04 | | Release date: | 2003-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of the Ribonuclease MC1 Mutants N71T and N71S in Complex with 5'-GMP: Structural Basis for Alterations in Substrate Specificity
Biochemistry, 42, 2003
|
|
1J1F
 
 | | Crystal structure of the RNase MC1 mutant N71T in complex with 5'-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, RIBONUCLEASE MC1 | | Authors: | Numata, T, Suzuki, A, Kakuta, Y, Kimura, K, Yao, M, Tanaka, I, Yoshida, Y, Ueda, T, Kimura, M. | | Deposit date: | 2002-12-03 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of the Ribonuclease MC1 Mutants N71T and N71S in Complex with 5'-GMP: Structural Basis for Alterations in Substrate Specificity
Biochemistry, 42, 2003
|
|
2L3T
 
 | | Solution structure of tandem SH2 domain from Spt6 | | Descriptor: | Transcription elongation factor SPT6 | | Authors: | Liu, J, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2010-09-22 | | Release date: | 2011-06-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the tandem SH2 domains from Spt6 and their binding to the phosphorylated RNA polymerase II C-terminal domain
To be Published
|
|
1BQT
 
 | | THREE-DIMENSIONAL STRUCTURE OF HUMAN INSULIN-LIKE GROWTH FACTOR-I (IGF-I) DETERMINED BY 1H-NMR AND DISTANCE GEOMETRY, 6 STRUCTURES | | Descriptor: | INSULIN-LIKE GROWTH FACTOR-I | | Authors: | Sato, A, Nishimura, S, Ohkubo, T, Kyogoku, Y, Koyama, S, Kobayashi, M, Yasuda, T, Kobayashi, Y. | | Deposit date: | 1998-08-18 | | Release date: | 1999-05-18 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of human insulin-like growth factor-I (IGF-I) determined by 1H-NMR and distance geometry.
Int.J.Pept.Protein Res., 41, 1993
|
|
1ISE
 
 | | Crystal structure of a mutant of ribosome recycling factor from Escherichia coli, Arg132Gly | | Descriptor: | Ribosome Recycling Factor | | Authors: | Nakano, H, Yoshida, T, Oka, S, Uchiyama, S, Nishina, K, Ohkubo, T, Kato, H, Yamagata, Y, Kobayashi, Y. | | Deposit date: | 2001-11-30 | | Release date: | 2003-10-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a mutant of ribosome recycling factor from Escherichia coli, Arg132Gly
To be Published
|
|
6JZW
 
 | |
6JZV
 
 | | Crystal structure of SufU from Bacillus subtilis | | Descriptor: | ZINC ION, Zinc-dependent sulfurtransferase SufU | | Authors: | Fujishiro, T, Takahashi, Y. | | Deposit date: | 2019-05-04 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cysteine-Persulfide Sulfane Sulfur-Ligated Zn Complex of Sulfur-Carrying SufU in the SufCDSUB System for Fe-S Cluster Biosynthesis.
Inorg.Chem., 2024
|
|
