2AIN
 
 | |
6L4P
 
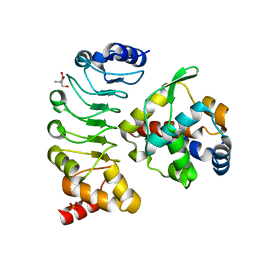 | | Crystal structure of the complex between the axonemal outer-arm dynein light chain-1 and microtubule binding domain of gamma heavy chain | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dynein light chain 1, axonemal, ... | | Authors: | Toda, A, Nishikawa, Y, Tanaka, H, Yagi, T, Kurisu, G. | | Deposit date: | 2019-10-19 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | The complex of outer-arm dynein light chain-1 and the microtubule-binding domain of the gamma heavy chain shows how axonemal dynein tunes ciliary beating.
J.Biol.Chem., 295, 2020
|
|
1BQT
 
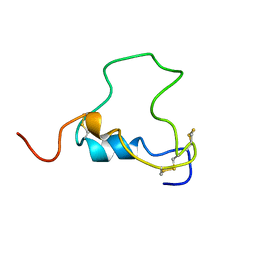 | | THREE-DIMENSIONAL STRUCTURE OF HUMAN INSULIN-LIKE GROWTH FACTOR-I (IGF-I) DETERMINED BY 1H-NMR AND DISTANCE GEOMETRY, 6 STRUCTURES | | Descriptor: | INSULIN-LIKE GROWTH FACTOR-I | | Authors: | Sato, A, Nishimura, S, Ohkubo, T, Kyogoku, Y, Koyama, S, Kobayashi, M, Yasuda, T, Kobayashi, Y. | | Deposit date: | 1998-08-18 | | Release date: | 1999-05-18 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of human insulin-like growth factor-I (IGF-I) determined by 1H-NMR and distance geometry.
Int.J.Pept.Protein Res., 41, 1993
|
|
4UX2
 
 | | Cryo-EM structure of antagonist-bound E2P gastric H,K-ATPase (SCH.E2. MgF) | | Descriptor: | POTASSIUM-TRANSPORTING ATPASE ALPHA CHAIN 1, POTASSIUM-TRANSPORTING ATPASE SUBUNIT BETA | | Authors: | Abe, K, Tani, K, Fujiyoshi, Y. | | Deposit date: | 2014-08-18 | | Release date: | 2014-09-17 | | Last modified: | 2014-11-12 | | Method: | ELECTRON CRYSTALLOGRAPHY (7 Å) | | Cite: | Systematic Comparison of Molecular Conformations of H+,K+-ATPase Reveals an Important Contribution of the A-M2 Linker for the Luminal Gating.
J.Biol.Chem., 289, 2014
|
|
1Z8U
 
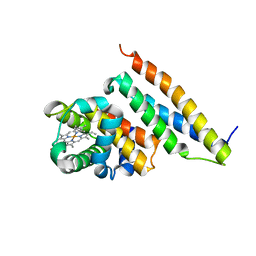 | | Crystal structure of oxidized alpha hemoglobin bound to AHSP | | Descriptor: | Alpha-hemoglobin stabilizing protein, Hemoglobin alpha chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Feng, L, Zhou, S, Gu, L, Gell, D.A, Mackay, J.P, Weiss, M.J, Gow, A.J, Shi, Y. | | Deposit date: | 2005-03-31 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of oxidized alpha-haemoglobin bound to AHSP reveals a protective mechanism for haem.
Nature, 435, 2005
|
|
1YOP
 
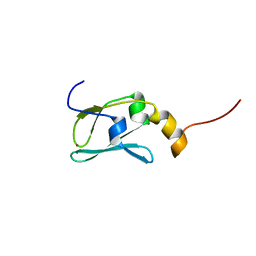 | | The solution structure of Kti11p | | Descriptor: | Kti11p, ZINC ION | | Authors: | Sun, J, Zhang, J, Wu, F, Xu, C, Li, S, Zhao, W, Wu, Z, Wu, J, Zhou, C.-Z, Shi, Y. | | Deposit date: | 2005-01-28 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Kti11p from Saccharomyces cerevisiae reveals a novel zinc-binding module.
Biochemistry, 44, 2005
|
|
2A5Y
 
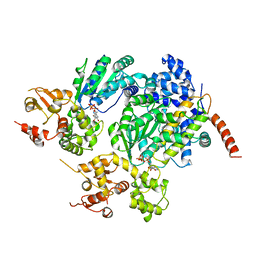 | | Structure of a CED-4/CED-9 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Apoptosis regulator ced-9, MAGNESIUM ION, ... | | Authors: | Yan, N, Liu, Q, Hao, Q, Gu, L, Shi, Y. | | Deposit date: | 2005-07-01 | | Release date: | 2005-10-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the CED-4-CED-9 complex provides insights into programmed cell death in Caenorhabditis elegans.
Nature, 437, 2005
|
|
8JVG
 
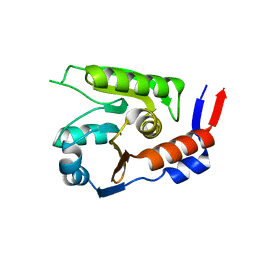 | | Crystal structure of dephospho-coenzyme A kinase | | Descriptor: | GTP-dependent dephospho-CoA kinase | | Authors: | Kita, A, Ishida, Y, Shimosaka, T, Michimori, Y, Makarova, K, Koonin, E, Atomi, H, Miki, K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of GTP-dependent dephospho-coenzyme A kinase from the hyperthermophilic archaeon, Thermococcus kodakarensis.
Proteins, 92, 2024
|
|
8JVC
 
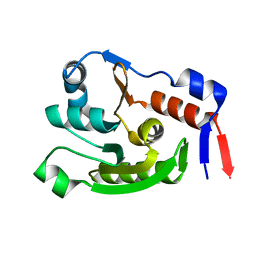 | | Crystal structure of dephospho-coenzyme A kinase | | Descriptor: | GTP-dependent dephospho-CoA kinase | | Authors: | Kita, A, Ishida, Y, Shimosaka, T, Michimori, Y, Makarova, K, Koonin, E, Atomi, H, Miki, K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of GTP-dependent dephospho-coenzyme A kinase from the hyperthermophilic archaeon, Thermococcus kodakarensis.
Proteins, 92, 2024
|
|
8JVF
 
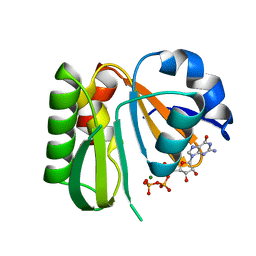 | | Crystal structure of dephospho-coenzyme A kinase | | Descriptor: | GTP-dependent dephospho-CoA kinase, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Kita, A, Ishida, Y, Shimosaka, T, Michimori, Y, Makarova, K, Koonin, E, Atomi, H, Miki, K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of GTP-dependent dephospho-coenzyme A kinase from the hyperthermophilic archaeon, Thermococcus kodakarensis.
Proteins, 92, 2024
|
|
5AZ2
 
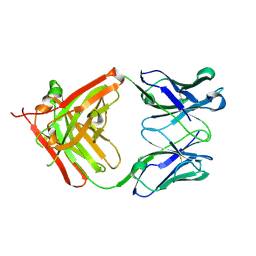 | | Crystal structure of the Fab fragment of 9E5, a murine monoclonal antibody specific for human epiregulin | | Descriptor: | anti-human epiregulin antibody 9E5 Fab heavy chain, anti-human epiregulin antibody 9E5 Fab light chain | | Authors: | Kado, Y, Mizohata, E, Nagatoishi, S, Iijima, M, Shinoda, K, Miyafusa, T, Nakayama, T, Yoshizumi, T, Sugiyama, A, Kawamura, T, Lee, Y.H, Matsumura, H, Doi, H, Fujitani, H, Kodama, T, Shibasaki, Y, Tsumoto, K, Inoue, T. | | Deposit date: | 2015-09-16 | | Release date: | 2015-12-09 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Epiregulin Recognition Mechanisms by Anti-epiregulin Antibody 9E5: STRUCTURAL, FUNCTIONAL, AND MOLECULAR DYNAMICS SIMULATION ANALYSES
J.Biol.Chem., 291, 2016
|
|
1AB9
 
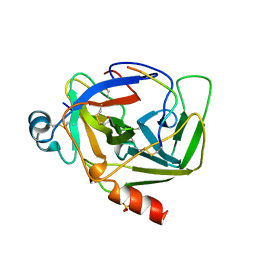 | | CRYSTAL STRUCTURE OF BOVINE GAMMA-CHYMOTRYPSIN | | Descriptor: | GAMMA-CHYMOTRYPSIN, PENTAPEPTIDE (TPGVY), SULFATE ION | | Authors: | Sugio, S, Kashima, A, Inoue, Y, Maeda, I, Nose, T, Shimohigashi, Y. | | Deposit date: | 1997-02-05 | | Release date: | 1997-08-20 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystal structure of a dipeptide-chymotrypsin complex in an inhibitory interaction.
Eur.J.Biochem., 255, 1998
|
|
5H0Q
 
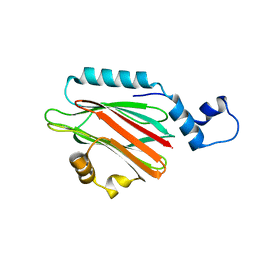 | | Crystal structure of lipid binding protein Nakanori at 1.5A | | Descriptor: | Lipid binding protein | | Authors: | Makino, A, Abe, M, Ishitsuka, R, Murate, M, Kishimoto, T, Sakai, S, Hullin-Matsuda, F, Shimada, Y, Inaba, T, Miyatake, H, Tanaka, H, Kurahashi, A, Pack, C.G, Kasai, R.S, Kubo, S, Schieber, N.L, Dohmae, N, Tochio, N, Hagiwara, K, Sasaki, Y, Aida, Y, Fujimori, F, Kigawa, T, Nishikori, K, Parton, R.G, Kusumi, A, Sako, Y, Anderluh, G, Yamashita, M, Kobayashi, T, Greimel, P, Kobayashi, T. | | Deposit date: | 2016-10-06 | | Release date: | 2016-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | A novel sphingomyelin/cholesterol domain-specific probe reveals the dynamics of the membrane domains during virus release and in Niemann-Pick type C
FASEB J., 31, 2017
|
|
4P79
 
 | | Crystal structure of mouse claudin-15 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Claudin-15 | | Authors: | Suzuki, H, Nishizawa, T, Tani, K, Yamazaki, Y, Tamura, A, Ishitani, R, Dohmae, N, Tsukita, S, Nureki, O, Fujiyoshi, Y. | | Deposit date: | 2014-03-26 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a claudin provides insight into the architecture of tight junctions.
Science, 344, 2014
|
|
1UG0
 
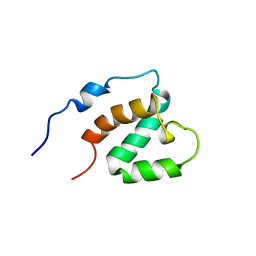 | | Solution structure of SURP domain in BAB30904 | | Descriptor: | splicing factor 4 | | Authors: | He, F, Muto, Y, Ushikoshi, R, Koshiba, S, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-11 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SURP domain in BAB30904
To be Published
|
|
1V9J
 
 | | Solution structure of a BolA-like protein from Mus musculus | | Descriptor: | BolA-like protein RIKEN cDNA 1110025L05 | | Authors: | Kasai, T, Inoue, M, Koshiba, S, Yabuki, T, Aoki, M, Nunokawa, E, Seki, E, Matsuda, T, Matsuda, N, Tomo, Y, Shirouzu, M, Terada, T, Obayashi, N, Hamana, H, Shinya, N, Tatsuguchi, A, Yasuda, S, Yoshida, M, Hirota, H, Matsuo, Y, Tani, K, Suzuki, H, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-26 | | Release date: | 2004-02-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a BolA-like protein from Mus musculus
Protein Sci., 13, 2004
|
|
1AT9
 
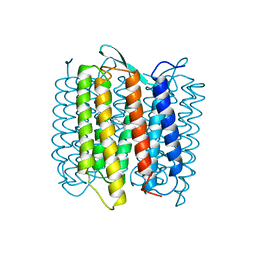 | | STRUCTURE OF BACTERIORHODOPSIN AT 3.0 ANGSTROM DETERMINED BY ELECTRON CRYSTALLOGRAPHY | | Descriptor: | BACTERIORHODOPSIN, RETINAL | | Authors: | Kimura, Y, Vassylyev, D.G, Miyazawa, A, Kidera, A, Matsushima, M, Mitsuoka, K, Murata, K, Hirai, T, Fujiyoshi, Y. | | Deposit date: | 1997-08-20 | | Release date: | 1998-09-16 | | Last modified: | 2024-06-05 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.8 Å) | | Cite: | Surface of bacteriorhodopsin revealed by high-resolution electron crystallography.
Nature, 389, 1997
|
|
4WY3
 
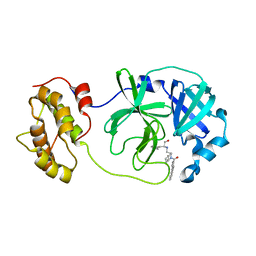 | | Structure of SARS-3CL protease complex with a phenylbenzoyl (R,S)-N-decalin type inhibitor | | Descriptor: | (2S)-2-({[(3R,4aS,8aR)-2-(biphenyl-4-ylcarbonyl)decahydroisoquinolin-3-yl]methyl}amino)-3-(1H-imidazol-5-yl)propanal, 3C-like proteinase | | Authors: | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2014-11-15 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors.
Bioorg.Med.Chem., 23, 2015
|
|
4DVY
 
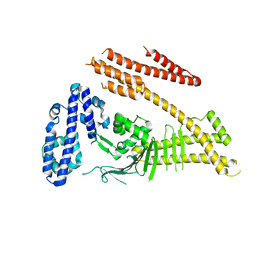 | | Crystal structure of the Helicobacter pylori CagA oncoprotein | | Descriptor: | Cytotoxicity-associated immunodominant antigen | | Authors: | Hayashi, T, Senda, M, Morohashi, H, Higashi, H, Horio, M, Kashiba, Y, Nagase, L, Sasaya, D, Shimizu, T, Venugopalan, N, Kumeta, H, Noda, N, Inagaki, F, Senda, T, Hatakeyama, M. | | Deposit date: | 2012-02-23 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Tertiary Structure-Function Analysis Reveals the Pathogenic Signaling Potentiation Mechanism of Helicobacter pylori Oncogenic Effector CagA
Cell Host Microbe, 12, 2012
|
|
7Q5X
 
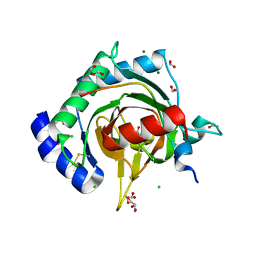 | | HIF PROLYL HYDROXYLASE 2 (PHD2/EGLN1) IN COMPLEX WITH 2-OXOGLUTARATE (2OG) AND HIF-2 ALPHA CODD (523-542) | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Figg Jr, W.D, McDonough, M.A, Chowdhury, R, Nakashima, Y, Schofield, C.J. | | Deposit date: | 2021-11-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural basis for binding of the renal carcinoma target hypoxia-inducible factor 2 alpha to prolyl hydroxylase domain 2.
Proteins, 91, 2023
|
|
1AFQ
 
 | | CRYSTAL STRUCTURE OF BOVINE GAMMA-CHYMOTRYPSIN COMPLEXED WITH A SYNTHETIC INHIBITOR | | Descriptor: | BOVINE GAMMA-CHYMOTRYPSIN, D-leucyl-N-(4-fluorobenzyl)-L-phenylalaninamide, SULFATE ION | | Authors: | Sugio, S, Kashima, A, Inoue, Y, Maeda, I, Nose, T, Shimohigashi, Y. | | Deposit date: | 1997-03-12 | | Release date: | 1997-09-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of a dipeptide-chymotrypsin complex in an inhibitory interaction.
Eur.J.Biochem., 255, 1998
|
|
4DVZ
 
 | | Crystal structure of the Helicobacter pylori CagA oncoprotein | | Descriptor: | Cytotoxicity-associated immunodominant antigen | | Authors: | Hayashi, T, Senda, M, Morohashi, H, Higashi, H, Horio, M, Kashiba, Y, Nagase, L, Sasaya, D, Shimizu, T, Venugopalan, N, Kumeta, H, Noda, N, Inagaki, F, Senda, T, Hatakeyama, M. | | Deposit date: | 2012-02-23 | | Release date: | 2012-07-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Tertiary structure-function analysis reveals the pathogenic signaling potentiation mechanism of Helicobacter pylori oncogenic effector CagA
Cell Host Microbe, 12, 2012
|
|
1BA5
 
 | | DNA-BINDING DOMAIN OF HUMAN TELOMERIC PROTEIN, HTRF1, NMR, 18 STRUCTURES | | Descriptor: | HTRF1 | | Authors: | Nishikawa, T, Nagadoi, A, Yoshimura, S, Aimoto, S, Nishimura, Y. | | Deposit date: | 1998-04-22 | | Release date: | 1999-04-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of human telomeric protein, hTRF1.
Structure, 6, 1998
|
|
5LAH
 
 | | NMR structure of the sea anemone peptide tau-AnmTx Ueq 12-1 with an uncommon fold | | Descriptor: | tau-AnmTx Ueq 12-1 | | Authors: | Mineev, K.S, Arseniev, A.S, Andreev, Y.A, Kozlov, S.A, Logashina, Y.A. | | Deposit date: | 2016-06-14 | | Release date: | 2017-05-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | New Disulfide-Stabilized Fold Provides Sea Anemone Peptide to Exhibit Both Antimicrobial and TRPA1 Potentiating Properties
Toxins, 9, 2017
|
|
3FHP
 
 | | A neutron crystallographic analysis of a porcine 2Zn insulin at 2.0 A resolution | | Descriptor: | Insulin, ZINC ION | | Authors: | Iwai, W, Kurihara, K, Yamada, T, Kobayashi, Y, Ohnishi, Y, Tanaka, I, Takahashi, H, Niimura, N. | | Deposit date: | 2008-12-09 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | NEUTRON DIFFRACTION (2 Å) | | Cite: | A neutron crystallographic analysis of T6 porcine insulin at 2.1 A resolution
Acta Crystallogr.,Sect.D, 65, 2009
|
|
