3VX7
 
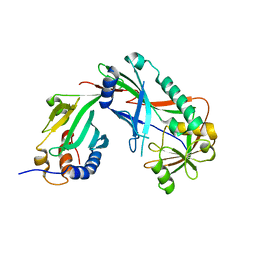 | | Crystal structure of Kluyveromyces marxianus Atg7NTD-Atg10 complex | | 分子名称: | E1, E2 | | 著者 | Yamaguchi, M, Matoba, K, Sawada, R, Fujioka, Y, Nakatogawa, H, Yamamoto, H, Kobashigawa, Y, Hoshida, H, Akada, R, Ohsumi, Y, Noda, N.N, Inagaki, F. | | 登録日 | 2012-09-11 | | 公開日 | 2012-11-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7.
Nat.Struct.Mol.Biol., 19, 2012
|
|
7LLQ
 
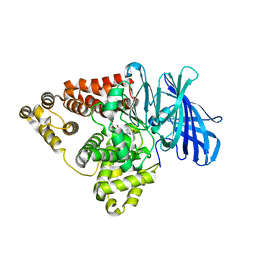 | | Substrate-dependent divergence of leukotriene A4 hydrolase aminopeptidase activity | | 分子名称: | 1-benzyl-4-methoxybenzene, 1-{4-oxo-4-[(2S)-pyrrolidin-2-yl]butanoyl}-L-proline, Leukotriene A-4 hydrolase, ... | | 著者 | Lee, K.H, Lee, S.H, Shim, Y.M, Paige, M, Noble, S.M. | | 登録日 | 2021-02-04 | | 公開日 | 2022-06-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Substrate-dependent modulation of the leukotriene A 4 hydrolase aminopeptidase activity and effect in a murine model of acute lung inflammation.
Sci Rep, 12, 2022
|
|
1J05
 
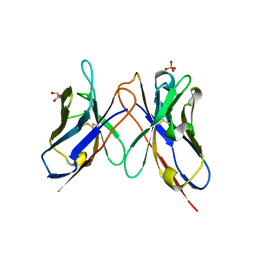 | | The crystal structure of anti-carcinoembryonic antigen monoclonal antibody T84.66 Fv fragment | | 分子名称: | GLYCEROL, PHOSPHATE ION, anti-CEA mAb T84.66, ... | | 著者 | Kondo, H, Nishimura, Y, Shiroishi, M, Asano, R, Noro, N, Tsumoto, K, Kumagai, I. | | 登録日 | 2002-11-01 | | 公開日 | 2003-12-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The crystal structure of anti-carcinoembryonic antigen monoclonal antibody T84.66 Fv fragment
To be Published
|
|
1J1N
 
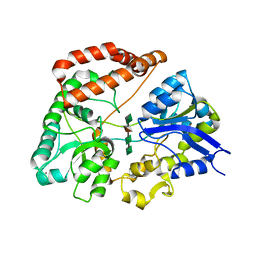 | | Structure Analysis of AlgQ2, A Macromolecule(Alginate)-Binding Periplasmic Protein Of Sphingomonas Sp. A1., Complexed with an Alginate Tetrasaccharide | | 分子名称: | AlgQ2, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid | | 著者 | Momma, K, Mikami, B, Mishima, Y, Hashimoto, W, Murata, K. | | 登録日 | 2002-12-11 | | 公開日 | 2003-06-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of AlgQ2, a macromolecule (alginate)-binding protein of Sphingomonas sp. A1, complexed with an alginate tetrasaccharide at 1.6-A resolution
J.BIOL.CHEM., 278, 2003
|
|
1IPI
 
 | |
5Y5H
 
 | | SF-ROX structure of cytochrome P450nor (NO-bound state) determined at SACLA | | 分子名称: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | 著者 | Tosha, T, Nomura, T, Nishida, T, Yamagiwa, R, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Yamashita, A, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Sugimoto, H, Shiro, Y, Kubo, M. | | 登録日 | 2017-08-09 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
1IQZ
 
 | | OXIDIZED [4Fe-4S] FERREDOXIN FROM BACILLUS THERMOPROTEOLYTICUS (FORM I) | | 分子名称: | Ferredoxin, IRON/SULFUR CLUSTER, SULFATE ION | | 著者 | Fukuyama, K, Okada, T, Kakuta, Y, Takahashi, Y. | | 登録日 | 2001-08-30 | | 公開日 | 2002-02-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (0.92 Å) | | 主引用文献 | Atomic resolution structures of oxidized [4Fe-4S] ferredoxin from Bacillus thermoproteolyticus in two crystal forms: systematic distortion of [4Fe-4S] cluster in the protein.
J.Mol.Biol., 315, 2002
|
|
1IR0
 
 | | OXIDIZED [4Fe-4S] FERREDOXIN FROM BACILLUS THERMOPROTEOLYTICUS (FORM II) | | 分子名称: | Ferredoxin, IRON/SULFUR CLUSTER, SULFATE ION | | 著者 | Fukuyama, K, Okada, T, Kakuta, Y, Takahashi, Y. | | 登録日 | 2001-08-30 | | 公開日 | 2002-02-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Atomic resolution structures of oxidized [4Fe-4S] ferredoxin from Bacillus thermoproteolyticus in two crystal forms: systematic distortion of [4Fe-4S] cluster in the protein.
J.Mol.Biol., 315, 2002
|
|
1ITY
 
 | | Solution structure of the DNA binding domain of human TRF1 | | 分子名称: | TRF1 | | 著者 | Nishikawa, T, Okamura, H, Nagadoi, A, Konig, P, Rhodes, D, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2002-02-15 | | 公開日 | 2002-03-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a telomeric DNA complex of human TRF1
Structure, 9, 2001
|
|
7WY4
 
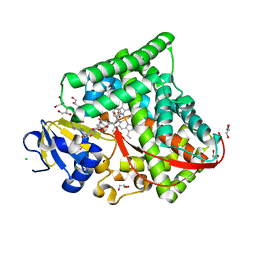 | | Structure of the CYP102A1 F87A Haem Domain with N-Enanthyl-L-Prolyl-L-Phenylalanine in complex with Styrene | | 分子名称: | (2S)-2-[[(2S)-1-heptylpyrrolidin-2-yl]carbonylamino]-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, CHLORIDE ION, ... | | 著者 | Suzuki, K, Stanfield, J.K, Shisaka, Y, Omura, K, Kasai, C, Sugimoto, H, Shoji, O. | | 登録日 | 2022-02-15 | | 公開日 | 2023-01-04 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | A Compound I Mimic Reveals the Transient Active Species of a Cytochrome P450 Enzyme: Insight into the Stereoselectivity of P450-Catalysed Oxidations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7WY3
 
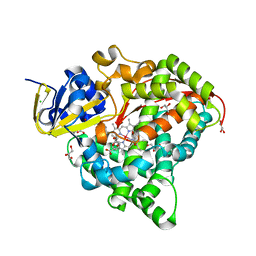 | | Structure of the Oxomolybdenum Mesoporphyrin IX-Reconstituted CYP102A1 F87V Mutant Haem Domain with N-(5-Cyclohexyl)valeroyl-L-Phenylalanine in complex with Styrene | | 分子名称: | (2~{S})-2-(5-cyclohexylpentanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, CHLORIDE ION, ... | | 著者 | Suzuki, K, Stanfield, J.K, Shisaka, Y, Omura, K, Kasai, C, Sugimoto, H, Shoji, O. | | 登録日 | 2022-02-15 | | 公開日 | 2023-01-04 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A Compound I Mimic Reveals the Transient Active Species of a Cytochrome P450 Enzyme: Insight into the Stereoselectivity of P450-Catalysed Oxidations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7WY2
 
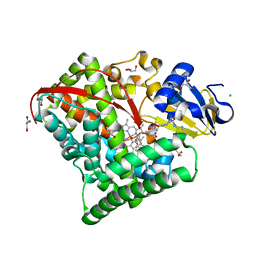 | | Structure of the Oxomolybdenum Mesoporphyrin IX-Reconstituted CYP102A1 F87A Mutant Haem Domain with N-Enanthyl-L-Prolyl-L-Phenylalanine in complex with Styrene | | 分子名称: | (2S)-2-[[(2S)-1-heptylpyrrolidin-2-yl]carbonylamino]-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, CHLORIDE ION, ... | | 著者 | Suzuki, K, Stanfield, J.K, Shisaka, Y, Omura, K, Kasai, C, Sugimoto, H, Shoji, O. | | 登録日 | 2022-02-15 | | 公開日 | 2023-01-04 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | A Compound I Mimic Reveals the Transient Active Species of a Cytochrome P450 Enzyme: Insight into the Stereoselectivity of P450-Catalysed Oxidations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
1IO7
 
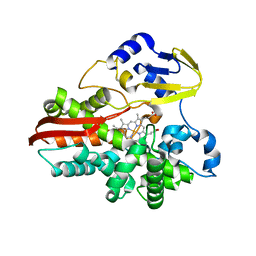 | | THERMOPHILIC CYTOCHROME P450 (CYP119) FROM SULFOLOBUS SOLFATARICUS: HIGH RESOLUTION STRUCTURAL ORIGIN OF ITS THERMOSTABILITY AND FUNCTIONAL PROPERTIES | | 分子名称: | CYTOCHROME P450 CYP119, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Park, S.-Y, Yamane, K, Adachi, S, Shiro, Y, Sligar, S.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2001-02-08 | | 公開日 | 2001-02-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Thermophilic cytochrome P450 (CYP119) from Sulfolobus solfataricus: high resolution structure and functional properties.
J.Inorg.Biochem., 91, 2002
|
|
2RNQ
 
 | |
8J9F
 
 | | Structure of STG-hydrolyzing beta-glucosidase 1 (PSTG1) | | 分子名称: | Beta-glucosidase, GLYCEROL | | 著者 | Yanai, T, Imaizumi, R, Takahashi, Y, Katsumura, E, Yamamoto, M, Nakayama, T, Yamashita, S, Takeshita, K, Sakai, N, Matsuura, H. | | 登録日 | 2023-05-03 | | 公開日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structural insights into a bacterial beta-glucosidase that is capable of degrading sesaminol triglucoside to produce sesaminol
To be published
|
|
1IYK
 
 | | Crystal structure of candida albicans N-myristoyltransferase with myristoyl-COA and peptidic inhibitor | | 分子名称: | MYRISTOYL-COA:PROTEIN N-MYRISTOYLTRANSFERASE, TETRADECANOYL-COA, [CYCLOHEXYLETHYL]-[[[[4-[2-METHYL-1-IMIDAZOLYL-BUTYL]PHENYL]ACETYL]-SERYL]-LYSINYL]-AMINE | | 著者 | Sogabe, S, Fukami, T.A, Morikami, K, Shiratori, Y, Aoki, Y, D'Arcy, A, Winkler, F.K, Banner, D.W, Ohtsuka, T. | | 登録日 | 2002-08-29 | | 公開日 | 2002-12-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structures of Candida albicans N-Myristoyltransferase with Two Distinct Inhibitors
CHEM.BIOL., 9, 2002
|
|
1IW1
 
 | | Crystal structure of a heme oxygenase (HmuO) from Corynebacterium diphtheriae complexed with heme in the ferrous state | | 分子名称: | Heme oxygenase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | 著者 | Hirotsu, S, Unno, M, Chu, G.C, Lee, D.S, Park, S.Y, Shiro, Y, Ikeda-Saito, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2002-04-04 | | 公開日 | 2003-04-04 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The crystal structures of the ferric and ferrous forms of the heme complex of HmuO, a heme oxygenase of Corynebacterium diphtheriae.
J.Biol.Chem., 279, 2004
|
|
1IYL
 
 | | Crystal Structure of Candida albicans N-myristoyltransferase with Non-peptidic Inhibitor | | 分子名称: | (1-METHYL-1H-IMIDAZOL-2-YL)-(3-METHYL-4-{3-[(PYRIDIN-3-YLMETHYL)-AMINO]-PROPOXY}-BENZOFURAN-2-YL)-METHANONE, Myristoyl-CoA:Protein N-Myristoyltransferase | | 著者 | Sogabe, S, Fukami, T.A, Morikami, K, Shiratori, Y, Aoki, Y, D'Arcy, A, Winkler, F.K, Banner, D.W, Ohtsuka, T. | | 登録日 | 2002-08-29 | | 公開日 | 2002-12-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Crystal Structures of Candida albicans N-Myristoyltransferase with Two Distinct Inhibitors
CHEM.BIOL., 9, 2002
|
|
8J7R
 
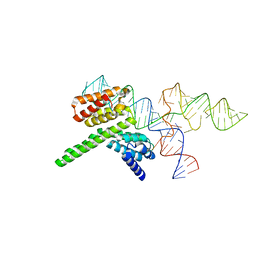 | | Cryo-EM structure of the J-K-St region of EMCV IRES in complex with eIF4G-HEAT1 and eIF4A (J-K-St/eIF4G focused) | | 分子名称: | Eukaryotic translation initiation factor 4 gamma 1, IRES RNA (J-K-St), MAGNESIUM ION | | 著者 | Suzuki, H, Fujiyoshi, Y, Imai, S, Shimada, I. | | 登録日 | 2023-04-28 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-09-13 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Dynamically regulated two-site interaction of viral RNA to capture host translation initiation factor.
Nat Commun, 14, 2023
|
|
1J1G
 
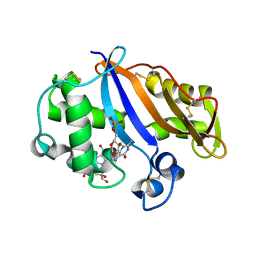 | | Crystal structure of the RNase MC1 mutant N71S in complex with 5'-GMP | | 分子名称: | GUANOSINE-5'-MONOPHOSPHATE, Ribonuclease MC1 | | 著者 | Numata, T, Suzuki, A, Kakuta, Y, Kimura, K, Yao, M, Tanaka, I, Yoshida, Y, Ueda, T, Kimura, M. | | 登録日 | 2002-12-04 | | 公開日 | 2003-05-20 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structures of the Ribonuclease MC1 Mutants N71T and N71S in Complex with 5'-GMP: Structural Basis for Alterations in Substrate Specificity
Biochemistry, 42, 2003
|
|
1J1F
 
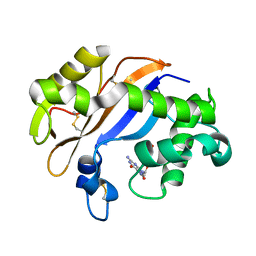 | | Crystal structure of the RNase MC1 mutant N71T in complex with 5'-GMP | | 分子名称: | GUANOSINE-5'-MONOPHOSPHATE, RIBONUCLEASE MC1 | | 著者 | Numata, T, Suzuki, A, Kakuta, Y, Kimura, K, Yao, M, Tanaka, I, Yoshida, Y, Ueda, T, Kimura, M. | | 登録日 | 2002-12-03 | | 公開日 | 2003-05-20 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structures of the Ribonuclease MC1 Mutants N71T and N71S in Complex with 5'-GMP: Structural Basis for Alterations in Substrate Specificity
Biochemistry, 42, 2003
|
|
7YH7
 
 | | SARS-CoV-2 spike in complex with neutralizing antibody NIV-8 (state 2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, ... | | 著者 | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | 登録日 | 2022-07-13 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
5Y5F
 
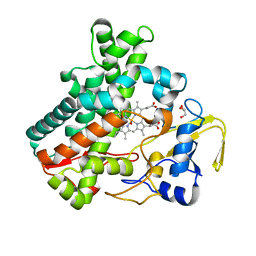 | | Structure of cytochrome P450nor in NO-bound state: damaged by low-dose (0.72 MGy) X-ray | | 分子名称: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | 著者 | Tosha, T, Nomura, T, Nishida, T, Ueno, G, Murakami, H, Yamashita, K, Hirata, K, Yamamoto, M, Ago, H, Sugimoto, H, Shiro, Y, Kubo, M. | | 登録日 | 2017-08-09 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5G
 
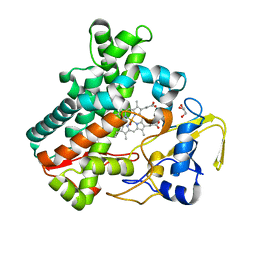 | | Structure of cytochrome P450nor in NO-bound state: damaged by high-dose (5.7 MGy) X-ray | | 分子名称: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | 著者 | Tosha, T, Nomura, T, Nishida, T, Ueno, G, Murakami, H, Yamashita, K, Hirata, K, Yamamoto, M, Ago, H, Sugimoto, H, Shiro, Y, Kubo, M. | | 登録日 | 2017-08-09 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
6FWF
 
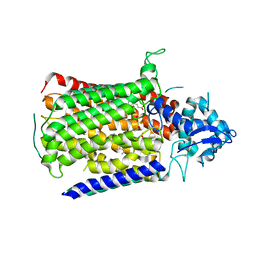 | | Low resolution structure of Neisseria meningitidis qNOR | | 分子名称: | CALCIUM ION, FE (III) ION, Nitric-oxide reductase, ... | | 著者 | Young, D, Antonyuk, S, Tosha, T, Hisano, T, Hasnain, S, Shiro, Y. | | 登録日 | 2018-03-06 | | 公開日 | 2018-03-14 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (4.2 Å) | | 主引用文献 | Characterization of the quinol-dependent nitric oxide reductase from the pathogen Neisseria meningitidis, an electrogenic enzyme.
Sci Rep, 8, 2018
|
|
