3VXG
 
 | | Crystal structure of conjugated polyketone reductase C2 from Candida Parapsilosis | | Descriptor: | Conjugated polyketone reductase C2 | | Authors: | Qin, H.-M, Yamamura, A, Miyakawa, T, Maruoka, S, Ohtsuka, J, Nagata, K, Kataoka, M, Shimizu, S, Tanokura, M. | | Deposit date: | 2012-09-13 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of conjugated polyketone reductase from Candida parapsilosis IFO 0708 reveals conformational changes for substrate recognition upon NADPH binding
Appl.Microbiol.Biotechnol., 98, 2014
|
|
4UBR
 
 | |
3VDQ
 
 | | Crystal structure of alcaligenes faecalis D-3-hydroxybutyrate dehydrogenase in complex with NAD(+) and acetate | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hoque, M.M, Shimizu, S, Hossain, M.T, Yamamoto, T, Suzuki, K, Takenaka, A. | | Deposit date: | 2012-01-06 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structures of Alcaligenes faecalis D-3-hydroxybutyrate dehydrogenase before and after NAD+ and acetate binding suggest a dynamical reaction mechanism as a member of the SDR family.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3VDR
 
 | | Crystal structure of D-3-hydroxybutyrate dehydrogenase, prepared in the presence of the substrate D-3-hydroxybutyrate and NAD(+) | | Descriptor: | (3R)-3-hydroxybutanoic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ACETOACETIC ACID, ... | | Authors: | Hoque, M.M, Shimizu, S, Juan, E.C.M, Sato, Y, Hossain, M.T, Yamamoto, T, Imamura, S, Amano, H, Suzuki, K, Sekiguchi, T, Tsunoda, M, Takenaka, A. | | Deposit date: | 2012-01-06 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of D-3-hydroxybutyrate dehydrogenase prepared in the presence of the substrate D-3-hydroxybutyrate and NAD+.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3WG6
 
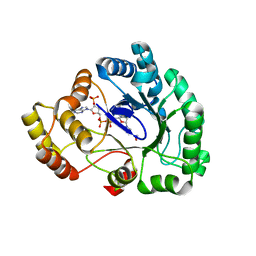 | | Crystal structure of conjugated polyketone reductase C1 from Candida parapsilosis complexed with NADPH | | Descriptor: | Conjugated polyketone reductase C1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Qin, H.-M, Yamamura, A, Miyakawa, T, Maruoka, S, Ohtsuka, J, Nagata, K, Kataoka, M, Shimizu, S, Tanokura, M. | | Deposit date: | 2013-07-28 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of conjugated polyketone reductase (CPR-C1) from Candida parapsilosis IFO 0708 complexed with NADPH.
Proteins, 81, 2013
|
|
8IJW
 
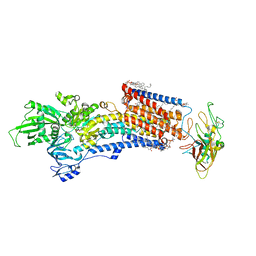 | | Cryo-EM structure of the gastric proton pump with bound DQ-06 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Abe, K, Yokoshima, S, Yoshimori, A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Deep learning driven de novo drug design based on gastric proton pump structures.
Commun Biol, 6, 2023
|
|
8IJX
 
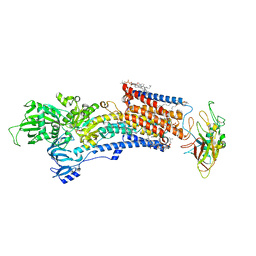 | | Cryo-EM structure of the gastric proton pump with bound DQ-18 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-[4-[(5-chloranyl-2-phenylmethoxy-phenyl)methoxy]phenyl]-N-methyl-methanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Abe, K, Yokoshima, S, Yoshimori, A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.08 Å) | | Cite: | Deep learning driven de novo drug design based on gastric proton pump structures.
Commun Biol, 6, 2023
|
|
8IJV
 
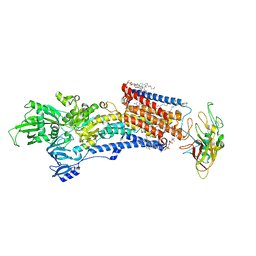 | | Cryo-EM structure of the gastric proton pump with bound DQ-02 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[[5-chloranyl-2-(4-chlorophenyl)phenyl]methoxy]-N-methyl-but-2-yn-1-amine, ... | | Authors: | Abe, K, Yokoshima, S, Yoshimori, A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Deep learning driven de novo drug design based on gastric proton pump structures.
Commun Biol, 6, 2023
|
|
7BKD
 
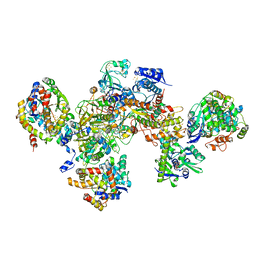 | | Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase complex from Methanospirillum hungatei (heterodislfide reductase core and mobile arm in conformational state 1, composite structure) | | Descriptor: | CoB--CoM heterodisulfide reductase iron-sulfur subunit A, CoB--CoM heterodisulfide reductase subunit B, CoB--CoM heterodisulfide reductase subunit C, ... | | Authors: | Pfeil-Gardiner, O, Watanabe, T, Shima, S, Murphy, B.J. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Three-megadalton complex of methanogenic electron-bifurcating and CO 2 -fixing enzymes.
Science, 373, 2021
|
|
7BKE
 
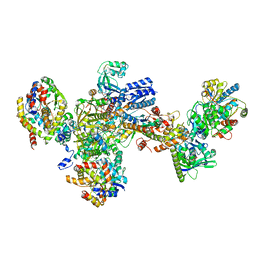 | | Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase complex from Methanospirillum hungatei (heterodisulfide reductase core and mobile arm in conformational state 2, composite structure) | | Descriptor: | CoB--CoM heterodisulfide reductase iron-sulfur subunit A, CoB--CoM heterodisulfide reductase subunit B, CoB--CoM heterodisulfide reductase subunit C, ... | | Authors: | Pfeil-Gardiner, O, Watanabe, T, Shima, S, Murphy, B.J. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Three-megadalton complex of methanogenic electron-bifurcating and CO 2 -fixing enzymes.
Science, 373, 2021
|
|
7BKB
 
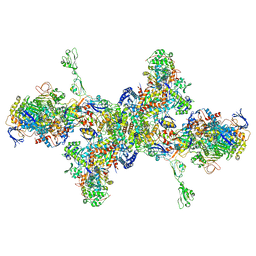 | | Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase complex from Methanospirillum hungatei (hexameric, composite structure) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CoB--CoM heterodisulfide reductase iron-sulfur subunit A, CoB--CoM heterodisulfide reductase subunit B, ... | | Authors: | Pfeil-Gardiner, O, Watanabe, T, Shima, S, Murphy, B.J. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Three-megadalton complex of methanogenic electron-bifurcating and CO 2 -fixing enzymes.
Science, 373, 2021
|
|
7BKC
 
 | | Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase complex from Methanospirillum hungatei (dimeric, composite structure) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CoB--CoM heterodisulfide reductase iron-sulfur subunit A, CoB--CoM heterodisulfide reductase subunit B, ... | | Authors: | Pfeil-Gardiner, O, Watanabe, T, Shima, S, Murphy, B.J. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Three-megadalton complex of methanogenic electron-bifurcating and CO 2 -fixing enzymes.
Science, 373, 2021
|
|
8JMN
 
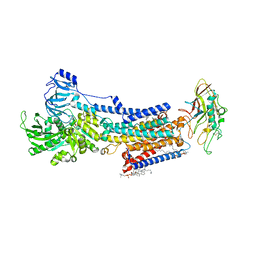 | | Cryo-EM structure of the gastric proton pump with bound DQ-21 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-[4-[[2-[(4-chlorophenyl)methoxy]phenyl]methoxy]phenyl]-N-methyl-methanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Abe, K, Yokoshima, S, Yoshimori, A. | | Deposit date: | 2023-06-05 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Deep learning driven de novo drug design based on gastric proton pump structures.
Commun Biol, 6, 2023
|
|
5LB9
 
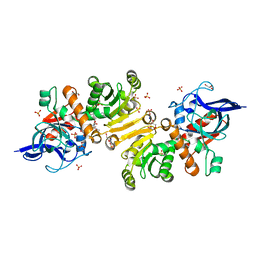 | | Structure of the T175V Etr1p mutant in the monoclinic form P21 | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADPH, B-specific] 1, mitochondrial, ... | | Authors: | Wagner, T, Rosenthal, R.G, Voegeli, B, Shima, S, Erb, T.J. | | Deposit date: | 2016-06-15 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A conserved threonine prevents self-intoxication of enoyl-thioester reductases.
Nat. Chem. Biol., 13, 2017
|
|
5AZE
 
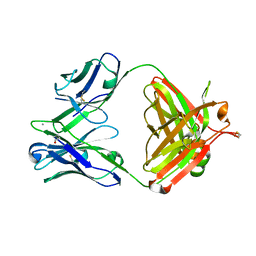 | | Fab fragment of calcium-dependent antigen binding antibody, 6RL#9 | | Descriptor: | 6RL#9 FAB HEAVY CHAIN, 6RL#9 FAB LIGHT CHAIN, CALCIUM ION | | Authors: | Kadono, S, Hironiwa, N, Ishii, S, Igawa, T, Hattori, K. | | Deposit date: | 2015-10-02 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Calcium-dependent antigen binding as a novel modality for antibody recycling by endosomal antigen dissociation
Mabs, 8, 2016
|
|
7XMA
 
 | | Crystal structure of Bovine heart cytochrome c oxidase, apo structure with DMSO | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Nishida, Y, Shinzawa-Itoh, K, Mizuno, N, Kumasaka, T, Yoshikawa, S, Tsukihara, T, Takashima, S, Shintani, Y. | | Deposit date: | 2022-04-25 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
7XMB
 
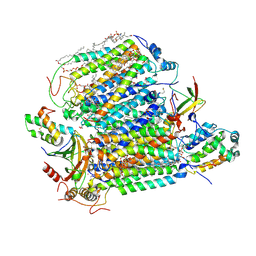 | | Crystal structure of Bovine heart cytochrome c oxidase, the structure complexed with an allosteric inhibitor T113 | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Nishida, Y, Shinzawa-Itoh, K, Mizuno, N, Kumasaka, T, Yoshikawa, S, Tsukihara, T, Shintani, Y, Takashima, S. | | Deposit date: | 2022-04-25 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
8JQQ
 
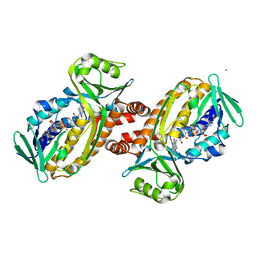 | | Protocatecuate hydroxylase from Xylophilus ampelinus C347T mutant | | Descriptor: | 4-hydroxybenzoate 3-monooxygenase (NAD(P)H), CALCIUM ION, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fukushima, R, Katsuki, N, Fushinobu, S, Takaya, N. | | Deposit date: | 2023-06-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Protocatechuate hydroxylase is a novel group A flavoprotein monooxygenase with a unique substrate recognition mechanism.
J.Biol.Chem., 300, 2023
|
|
8JQP
 
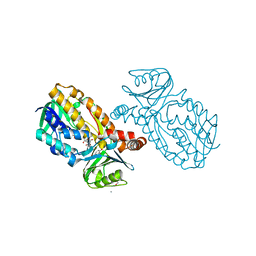 | | Protocatecuate hydroxylase from Xylophilus ampelinus complexed with 3,4-dihydroxybenzoate | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, 4-hydroxybenzoate 3-monooxygenase (NAD(P)H), CALCIUM ION, ... | | Authors: | Fukushima, R, Katsuki, N, Fushinobu, S, Takaya, N. | | Deposit date: | 2023-06-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Protocatechuate hydroxylase is a novel group A flavoprotein monooxygenase with a unique substrate recognition mechanism.
J.Biol.Chem., 300, 2023
|
|
8JQO
 
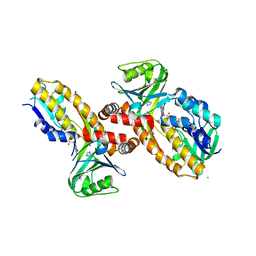 | | Protocatecuate hydroxylase from Xylophilus ampelinus complexed with imidazole | | Descriptor: | 4-hydroxybenzoate 3-monooxygenase (NAD(P)H), CALCIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Fukushima, R, Katsuki, N, Fushinobu, S, Takaya, N. | | Deposit date: | 2023-06-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Protocatechuate hydroxylase is a novel group A flavoprotein monooxygenase with a unique substrate recognition mechanism.
J.Biol.Chem., 300, 2023
|
|
5L8X
 
 | |
4PPC
 
 | |
4PPB
 
 | |
3GJJ
 
 | | crystal structure of a DNA duplex containing 7,8-dihydropyridol[2,3-d]pyrimidin-2-one | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, 5'-D(*CP*GP*CP*GP*AP*AP*TP*(B7C)P*CP*GP*CP*G)-3' | | Authors: | Takenaka, A, Juan, E.C.M, Shimizu, S, Haraguchi, T, Xiao, M, Kurose, T, Ohkubo, A, Sekine, M, Shibata, T, Millington, C.L, Williams, D.M. | | Deposit date: | 2009-03-09 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights into the stabilizing contributions of bicyclic cytosine analogues: crystal structures of DNA duplexes containing 7,8-dihydropyridol[2,3-d]pyrimidin-2-one
To be Published
|
|
4PPA
 
 | |
