2E1D
 
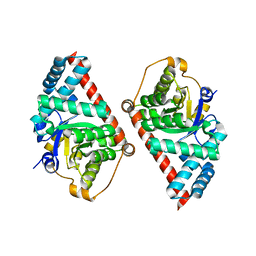 | | Crystal structure of mouse transaldolase | | Descriptor: | SULFITE ION, Transaldolase | | Authors: | Kishishita, S, Murayama, K, Chen, L, Liu, Z.J, Wang, B.C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-10-25 | | Release date: | 2007-11-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of mouse transaldolase
To be Published
|
|
3VOC
 
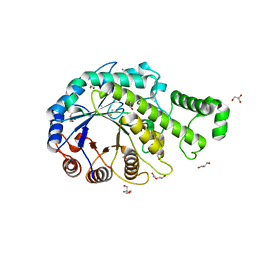 | | Crystal structure of the catalytic domain of beta-amylase from paenibacillus polymyxa | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta/alpha-amylase, ... | | Authors: | Nishimura, S, Fujioka, T, Nakaniwa, T, Tada, T. | | Deposit date: | 2012-01-21 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis by X-ray crystallography and small-angle scattering of the multi-domain beta-amylase from Paenibacillus polymyxa
To be Published
|
|
2ON9
 
 | |
3B1B
 
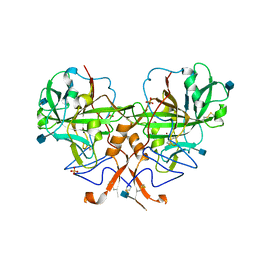 | |
2ONA
 
 | |
2ONV
 
 | |
2ONW
 
 | |
2QZL
 
 | | Crystal Structure of human Beta Secretase complexed with IXS | | Descriptor: | Beta-secretase 1, N-[(1S)-1-benzyl-2-{[(1S)-2-(isobutylamino)-1-methyl-2-oxoethyl]amino}ethyl]-N'-[(1R)-1-(4-fluorophenyl)ethyl]-5-[methyl(methylsulfonyl)amino]isophthalamide | | Authors: | Munshi, S. | | Deposit date: | 2007-08-16 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | BACE-1 inhibition by a series of psi[CH2NH] reduced amide isosteres
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2OAH
 
 | | Crystal Structure of Human Beta Secretase Complexed with inhibitor | | Descriptor: | Beta-secretase 1, N-[(1S,2S)-2-AMINO-1-(3-THIENYLMETHYL)HEXYL]-2-({[(1S,2S)-2-METHYLCYCLOPROPYL]METHYL}AMINO)-6-[METHYL(METHYLSULFONYL)AMINO]ISONICOTINAMIDE | | Authors: | Munshi, S. | | Deposit date: | 2006-12-15 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and SAR of isonicotinamide BACE-1 inhibitors that bind beta-secretase in a N-terminal 10s-loop down conformation.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2NTR
 
 | | Crystal structure of Human Bace-1 bound to inhibitor | | Descriptor: | (2R)-2-(5-{3-chloro-6-((2-methoxyethyl){[(1S,2S)-2-methylcyclopropyl]methyl}amino)-2-[methyl(methylsulfonyl)amino]pyrid in-4-yl}-1,3,4-oxadiazol-2-yl)-1-phenylpropan-2-amine, Beta-secretase 1 | | Authors: | Munshi, S. | | Deposit date: | 2006-11-08 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Beta-secretase (BACE-1) inhibitors: accounting for 10s loop flexibility using rigid active sites.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2P8H
 
 | | Crystal structure of human beta secretase complexed with inhibitor | | Descriptor: | Beta-secretase 1, N-{(1S,2S)-1-BENZYL-2-HYDROXY-2-[(4S)-1,2,2-TRIMETHYL-5-OXOIMIDAZOLIDIN-4-YL]ETHYL}-N'-[(1R)-1-(4-FLUOROPHENYL)ETHYL]-5-[METHYL(METHYLSULFONYL)AMINO]ISOPHTHALAMIDE | | Authors: | Munshi, S. | | Deposit date: | 2007-03-22 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and Synthesis of 2,3,5-Substituted Imidazolidin-4-one Inhibitors of BACE-1.
Chemmedchem, 2, 2007
|
|
1TZ1
 
 | | Solution structure of the PB1 domain of CDC24P (short form) | | Descriptor: | Cell division control protein 24 | | Authors: | Yoshinaga, S, Terasawa, H, Ogura, K, Noda, Y, Ito, T, Sumimoto, H, Inagaki, F. | | Deposit date: | 2004-07-09 | | Release date: | 2005-09-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PB1 domain of CDC24P (short form)
To be Published
|
|
1UK6
 
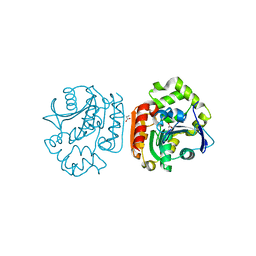 | | Crystal structure of a meta-cleavage product hydrolase (CumD) complexed with propionate | | Descriptor: | 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase, PROPANOIC ACID | | Authors: | Fushinobu, S, Jun, S.-Y, Hidaka, M, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | Deposit date: | 2003-08-19 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Series of Crystal Structures of a meta-Cleavage Product Hydrolase from Pseudomonas fluorescens IP01 (CumD) Complexed with Various Cleavage Products
BIOSCI.BIOTECHNOL.BIOCHEM., 69, 2005
|
|
1UK8
 
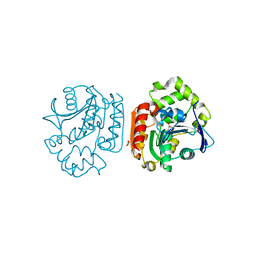 | | Crystal structure of a meta-cleavage product hydrolase (CumD) complexed with n-valerate | | Descriptor: | 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase, PENTANOIC ACID | | Authors: | Fushinobu, S, Jun, S.-Y, Hidaka, M, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | Deposit date: | 2003-08-19 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Series of Crystal Structures of a meta-Cleavage Product Hydrolase from Pseudomonas fluorescens IP01 (CumD) Complexed with Various Cleavage Products
BIOSCI.BIOTECHNOL.BIOCHEM., 69, 2005
|
|
1UK7
 
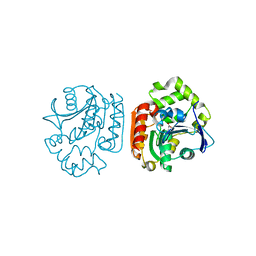 | | Crystal structure of a meta-cleavage product hydrolase (CumD) complexed with n-butyrate | | Descriptor: | 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase, butanoic acid | | Authors: | Fushinobu, S, Jun, S.-Y, Hidaka, M, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | Deposit date: | 2003-08-19 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Series of Crystal Structures of a meta-Cleavage Product Hydrolase from Pseudomonas fluorescens IP01 (CumD) Complexed with Various Cleavage Products
BIOSCI.BIOTECHNOL.BIOCHEM., 69, 2005
|
|
1UKA
 
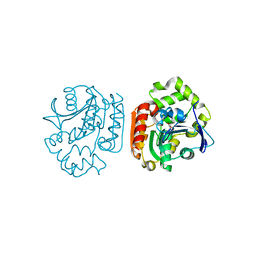 | | Crystal structure of a meta-cleavage product hydrolase (CumD) complexed with (S)-2-methylbutyrate | | Descriptor: | 2-METHYLBUTANOIC ACID, 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase | | Authors: | Fushinobu, S, Jun, S.-Y, Hidaka, M, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | Deposit date: | 2003-08-19 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Series of Crystal Structures of a meta-Cleavage Product Hydrolase from Pseudomonas fluorescens IP01 (CumD) Complexed with Various Cleavage Products
BIOSCI.BIOTECHNOL.BIOCHEM., 69, 2005
|
|
1UKB
 
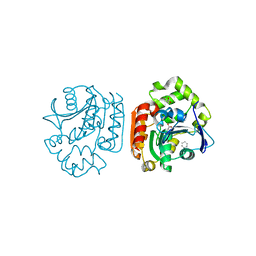 | | Crystal structure of a meta-cleavage product hydrolase (CumD) complexed with benzoate | | Descriptor: | 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase, BENZOIC ACID | | Authors: | Fushinobu, S, Jun, S.-Y, Hidaka, M, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | Deposit date: | 2003-08-19 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Series of Crystal Structures of a meta-Cleavage Product Hydrolase from Pseudomonas fluorescens IP01 (CumD) Complexed with Various Cleavage Products
BIOSCI.BIOTECHNOL.BIOCHEM., 69, 2005
|
|
1UK9
 
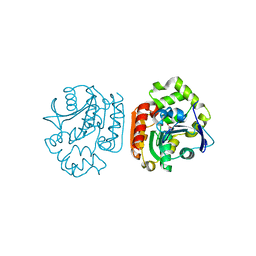 | | Crystal structure of a meta-cleavage product hydrolase (CumD) complexed with isovalerate | | Descriptor: | 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase, ISOVALERIC ACID | | Authors: | Fushinobu, S, Jun, S.-Y, Hidaka, M, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | Deposit date: | 2003-08-19 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Series of Crystal Structures of a meta-Cleavage Product Hydrolase from Pseudomonas fluorescens IP01 (CumD) Complexed with Various Cleavage Products
BIOSCI.BIOTECHNOL.BIOCHEM., 69, 2005
|
|
1TQF
 
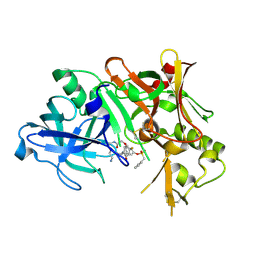 | | Crystal structure of human Beta secretase complexed with inhibitor | | Descriptor: | 3-{2-[(5-AMINOPENTYL)AMINO]-2-OXOETHOXY}-5-({[1-(4-FLUOROPHENYL)ETHYL]AMINO}CARBONYL)PHENYL PHENYLMETHANESULFONATE, Beta-secretase 1 | | Authors: | Munshi, S, Chen, Z, Kuo, L. | | Deposit date: | 2004-06-17 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of a small molecule nonpeptide active site beta-secretase inhibitor that displays a nontraditional binding mode for aspartyl proteases.
J.Med.Chem., 47, 2004
|
|
1WKK
 
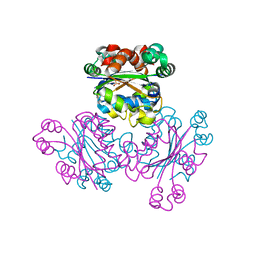 | |
1WKL
 
 | | Crystal Structure of Nucleoside Diphosphate Kinase from Thermus thermophilus HB8 in Complex with ATP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Takeishi, S, Nakagawa, N, Masui, R, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-06-01 | | Release date: | 2005-08-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Nucleoside Diphosphate Kinase from Thermus thermophilus HB8
To be Published
|
|
1WKJ
 
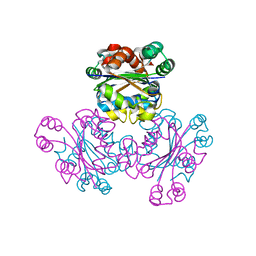 | |
2AC2
 
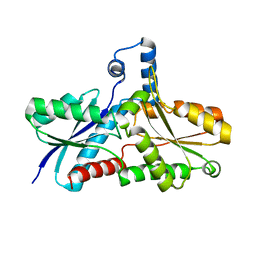 | | Crystal structure of the Tyr13Phe mutant variant of Bacillus subtilis Ferrochelatase with Zn(2+) bound at the active site | | Descriptor: | Ferrochelatase, ZINC ION | | Authors: | Shipovskov, S, Karlberg, T, Fodje, M, Hansson, M.D, Ferreira, G.C, Hansson, M, Reimann, C.T, Al-Karadaghi, S. | | Deposit date: | 2005-07-18 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Metallation of the Transition-state Inhibitor N-methyl Mesoporphyrin by Ferrochelatase: Implications for the Catalytic Reaction Mechanism.
J.Mol.Biol., 352, 2005
|
|
2AC4
 
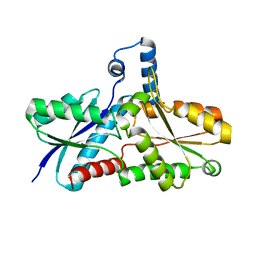 | | Crystal structure of the His183Cys mutant variant of Bacillus subtilis Ferrochelatase | | Descriptor: | Ferrochelatase | | Authors: | Shipovskov, S, Karlberg, T, Fodje, M, Hansson, M.D, Ferreira, G.C, Hansson, M, Reimann, C.T, Al-Karadaghi, S. | | Deposit date: | 2005-07-18 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Metallation of the Transition-state Inhibitor N-methyl Mesoporphyrin by Ferrochelatase: Implications for the Catalytic Reaction Mechanism.
J.Mol.Biol., 352, 2005
|
|
2YSD
 
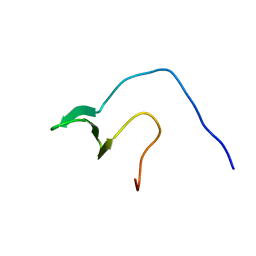 | | Solution structure of the first WW domain from the human membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1. MAGI-1 | | Descriptor: | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 | | Authors: | Ohnishi, S, Tochio, N, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first WW domain from the human membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1. MAGI-1
To be Published
|
|
