6K0I
 
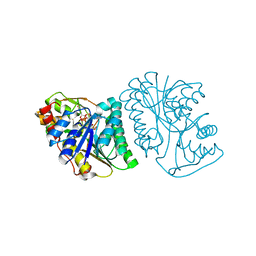 | | Crystal Structure of UDP-glucose 4-epimerase from Bifidobacterium longum in complex with NAD+ and UDP-Glc | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | 著者 | Nam, Y.-W, Nishimoto, M, Arakawa, T, Kitaoka, M, Fushinobu, S. | | 登録日 | 2019-05-06 | | 公開日 | 2019-08-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for broad substrate specificity of UDP-glucose 4-epimerase in the human milk oligosaccharide catabolic pathway of Bifidobacterium longum.
Sci Rep, 9, 2019
|
|
5GQF
 
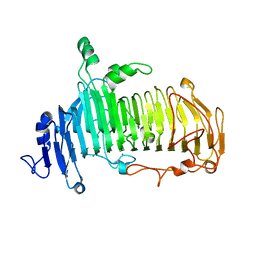 | | Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex | | 分子名称: | CALCIUM ION, Lacto-N-biosidase, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Yamada, C, Arakawa, T, Katayama, T, Fushinobu, S. | | 登録日 | 2016-08-07 | | 公開日 | 2017-04-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Molecular Insight into Evolution of Symbiosis between Breast-Fed Infants and a Member of the Human Gut Microbiome Bifidobacterium longum
Cell Chem Biol, 24, 2017
|
|
5D4T
 
 | |
6K0G
 
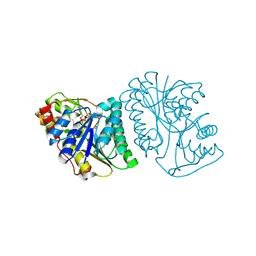 | | Crystal Structure of UDP-glucose 4-epimerase from Bifidobacterium longum in complex with NAD+ and UDP | | 分子名称: | MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase, ... | | 著者 | Nam, Y.-W, Nishimoto, M, Arakawa, T, Kitaoka, M, Fushinobu, S. | | 登録日 | 2019-05-06 | | 公開日 | 2019-08-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for broad substrate specificity of UDP-glucose 4-epimerase in the human milk oligosaccharide catabolic pathway of Bifidobacterium longum.
Sci Rep, 9, 2019
|
|
1CWP
 
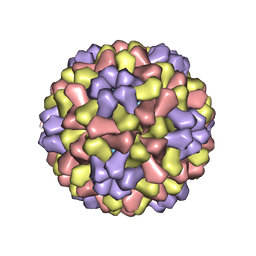 | | STRUCTURES OF THE NATIVE AND SWOLLEN FORMS OF COWPEA CHLOROTIC MOTTLE VIRUS DETERMINED BY X-RAY CRYSTALLOGRAPHY AND CRYO-ELECTRON MICROSCOPY | | 分子名称: | Coat protein, RNA (5'-R(*AP*U)-3'), RNA (5'-R(*AP*UP*AP*U)-3') | | 著者 | Speir, J.A, Johnson, J.E, Munshi, S, Wang, G, Timothy, S, Baker, T.S. | | 登録日 | 1995-05-22 | | 公開日 | 1995-05-22 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy.
Structure, 3, 1995
|
|
5D4V
 
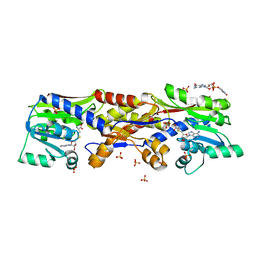 | | HcgC with SAH and a guanylylpyridinol (GP) derivative | | 分子名称: | 5'-O-[(R)-[(3,6-dimethyl-2-oxo-1,2-dihydropyridin-4-yl)oxy](hydroxy)phosphoryl]guanosine, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | 著者 | Fujishiro, T, Ermler, U, Shima, S. | | 登録日 | 2015-08-09 | | 公開日 | 2016-07-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Identification of HcgC as a SAM-Dependent Pyridinol Methyltransferase in [Fe]-Hydrogenase Cofactor Biosynthesis.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5D5T
 
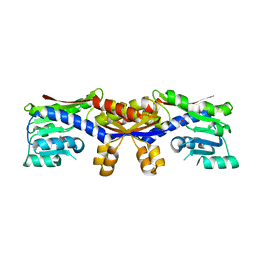 | |
5D5P
 
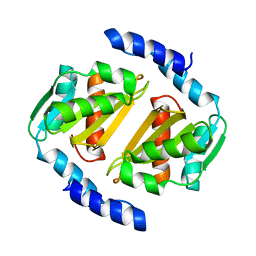 | |
1MBH
 
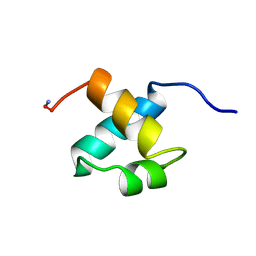 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 2 | | 分子名称: | C-MYB | | 著者 | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | 登録日 | 1995-05-19 | | 公開日 | 1995-09-15 | | 最終更新日 | 2024-06-05 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBF
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 1 | | 分子名称: | MYB PROTO-ONCOGENE PROTEIN | | 著者 | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | 登録日 | 1995-05-19 | | 公開日 | 1995-07-31 | | 最終更新日 | 2024-06-05 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
6KPL
 
 | | Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris in apo form | | 分子名称: | Chitinase, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL | | 著者 | Seki, H, Arakawa, T, Yamada, C, Takegawa, K, Fushinobu, S. | | 登録日 | 2019-08-15 | | 公開日 | 2019-10-02 | | 最終更新日 | 2019-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural basis for the specific cleavage of core-fucosylatedN-glycans by endo-beta-N-acetylglucosaminidase from the fungusCordyceps militaris.
J.Biol.Chem., 294, 2019
|
|
1MT4
 
 | | Structure of 23S ribosomal RNA hairpin 35 | | 分子名称: | 23S ribosomal Hairpin 35 | | 著者 | Lebars, I, Yoshizawa, S, Stenholm, A.R, Guittet, E, Douthwaite, S, Fourmy, D. | | 登録日 | 2002-09-20 | | 公開日 | 2003-01-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of 23S rRNA hairpin 35 and its interaction with the tylosin-resistance methyltransferase RlmAII
Embo J., 22, 2003
|
|
6KPM
 
 | | Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris in complex with L-fucose | | 分子名称: | Chitinase, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL, ... | | 著者 | Seki, H, Arakawa, T, Yamada, C, Takegawa, K, Fushinobu, S. | | 登録日 | 2019-08-15 | | 公開日 | 2019-10-02 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for the specific cleavage of core-fucosylatedN-glycans by endo-beta-N-acetylglucosaminidase from the fungusCordyceps militaris.
J.Biol.Chem., 294, 2019
|
|
1WQ3
 
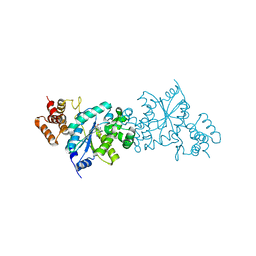 | | Escherichia coli tyrosyl-tRNA synthetase mutant complexed with 3-iodo-L-tyrosine | | 分子名称: | 3-IODO-TYROSINE, Tyrosyl-tRNA synthetase | | 著者 | Kobayashi, T, Sakamoto, K, Nureki, O, Takimura, T, Kamata, K, Sekine, R, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-09-20 | | 公開日 | 2005-01-25 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis of nonnatural amino acid recognition by an engineered aminoacyl-tRNA synthetase for genetic code expansion
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
6IOS
 
 | | The ligand binding domain of Mlp24 with proline | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | 著者 | Takahashi, Y, Sumita, K, Nishiyama, S, Kawagishi, I, Imada, K. | | 登録日 | 2018-10-31 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Calcium Ions Modulate Amino Acid Sensing of the Chemoreceptor Mlp24 ofVibrio cholerae.
J. Bacteriol., 201, 2019
|
|
1WQ4
 
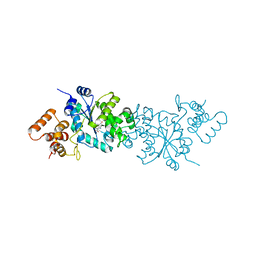 | | Escherichia coli tyrosyl-tRNA synthetase mutant complexed with L-tyrosine | | 分子名称: | TYROSINE, Tyrosyl-tRNA synthetase | | 著者 | Kobayashi, T, Sakamoto, K, Nureki, O, Takimura, T, Kamata, K, Sekine, R, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-09-20 | | 公開日 | 2005-01-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis of nonnatural amino acid recognition by an engineered aminoacyl-tRNA synthetase for genetic code expansion
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2RQJ
 
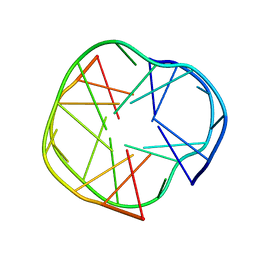 | | Quadruplex structure of an RNA aptamer against bovine prion protein | | 分子名称: | RNA (5'-R(*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*GP*A)-3') | | 著者 | Katahira, M, Mashima, T, Matsugami, A, Nishikawa, F, Nishikawa, S. | | 登録日 | 2009-07-18 | | 公開日 | 2009-11-17 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Unique quadruplex structure and interaction of an RNA aptamer against bovine prion protein
Nucleic Acids Res., 37, 2009
|
|
6IOP
 
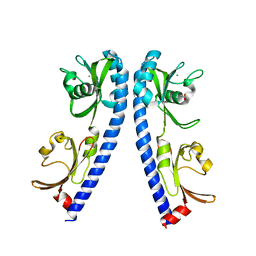 | | The ligand binding domain of Mlp24 | | 分子名称: | ACETATE ION, ALANINE, CALCIUM ION, ... | | 著者 | Sumita, K, Takahashi, Y, Nishiyama, S, Kawagishi, I, Imada, K. | | 登録日 | 2018-10-31 | | 公開日 | 2019-03-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Calcium Ions Modulate Amino Acid Sensing of the Chemoreceptor Mlp24 ofVibrio cholerae.
J. Bacteriol., 201, 2019
|
|
6IOV
 
 | | The ligand binding domain of Mlp37 with arginine | | 分子名称: | ARGININE, Methyl-accepting chemotaxis (MCP) signaling domain protein | | 著者 | Takahashi, Y, Sumita, K, Nishiyama, S, Kawagishi, I, Imada, K. | | 登録日 | 2018-10-31 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.351 Å) | | 主引用文献 | Structural basis of the binding affinity of chemoreceptors Mlp24p and Mlp37p for various amino acids.
Biochem.Biophys.Res.Commun., 523, 2020
|
|
2RSK
 
 | | RNA aptamer against prion protein in complex with the partial binding peptide | | 分子名称: | RNA (5'-R(*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*GP*A)-3'), partial binding peptide of Major prion protein | | 著者 | Mashima, T, Nishikawa, F, Kamatari, Y.O, Fujiwara, H, Nishikawa, S, Kuwata, K, Katahira, M. | | 登録日 | 2012-03-08 | | 公開日 | 2013-02-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Anti-prion activity of an RNA aptamer and its structural basis
Nucleic Acids Res., 41, 2013
|
|
6IOQ
 
 | | The ligand binding domain of Mlp24 with glycine | | 分子名称: | CALCIUM ION, GLYCINE, Methyl-accepting chemotaxis protein | | 著者 | Takahashi, Y, Sumita, K, Nishiyama, S, Kawagishi, I, Imada, K. | | 登録日 | 2018-10-31 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.143 Å) | | 主引用文献 | Calcium Ions Modulate Amino Acid Sensing of the Chemoreceptor Mlp24 ofVibrio cholerae.
J. Bacteriol., 201, 2019
|
|
6IOU
 
 | | The ligand binding domain of Mlp24 with serine | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Methyl-accepting chemotaxis protein, ... | | 著者 | Takahashi, Y, Sumita, K, Nishiyama, S, Kawagishi, I, Imada, K. | | 登録日 | 2018-10-31 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Calcium Ions Modulate Amino Acid Sensing of the Chemoreceptor Mlp24 ofVibrio cholerae.
J. Bacteriol., 201, 2019
|
|
6M5A
 
 | | Crystal structure of GH121 beta-L-arabinobiosidase HypBA2 from Bifidobacterium longum | | 分子名称: | 1,2-ETHANEDIOL, Beta-L-arabinobiosidase, CALCIUM ION, ... | | 著者 | Saito, K, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | 登録日 | 2020-03-10 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of beta-L-arabinobiosidase belonging to glycoside hydrolase family 121.
Plos One, 15, 2020
|
|
1X8X
 
 | | Tyrosyl t-RNA Synthetase from E.coli Complexed with Tyrosine | | 分子名称: | SULFATE ION, TYROSINE, Tyrosyl-tRNA synthetase | | 著者 | Kobayashi, T, Takimura, T, Sekine, R, Kelly, V.P, Kamata, K, Sakamoto, K, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-08-19 | | 公開日 | 2005-01-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Snapshots of the KMSKS Loop Rearrangement for Amino Acid Activation by Bacterial Tyrosyl-tRNA Synthetase
J.MOL.BIOL., 346, 2005
|
|
5YGY
 
 | | Crystal Structure of BACE1 in complex with (S)-N-(3-(2-amino-6-(fluoromethyl)-4 -methyl-4H-1,3-oxazin-4-yl)-4-fluorophenyl)-5-cyanopicolinamide | | 分子名称: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | 著者 | Fuchino, K, Mitsuoka, Y, Masui, M, Kurose, N, Yoshida, S, Komano, K, Yamamoto, T, Ogawa, M, Unemura, C, Hosono, M, Ito, H, Sakaguchi, G, Ando, S, Ohnishi, S, Kido, Y, Fukushima, T, Miyajima, H, Hiroyama, S, Koyabu, K, Dhuyvetter, D, Borghys, H, Gijsen, H, Yamano, Y, Iso, Y, Kusakabe, K. | | 登録日 | 2017-09-27 | | 公開日 | 2018-05-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Rational Design of Novel 1,3-Oxazine Based beta-Secretase (BACE1) Inhibitors: Incorporation of a Double Bond To Reduce P-gp Efflux Leading to Robust A beta Reduction in the Brain
J. Med. Chem., 61, 2018
|
|
