2MBC
 
 | | Solution Structure of human holo-PRL-3 in complex with vanadate | | Descriptor: | Protein tyrosine phosphatase type IVA 3 | | Authors: | Jeong, K, Kang, D, Kim, J, Shin, S, Jin, B, Lee, C, Kim, E, Jeon, Y.H, Kim, Y. | | Deposit date: | 2013-07-29 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and backbone dynamics of vanadate-bound PRL-3: comparison of 15N nuclear magnetic resonance relaxation profiles of free and vanadate-bound PRL-3.
Biochemistry, 53, 2014
|
|
3WP8
 
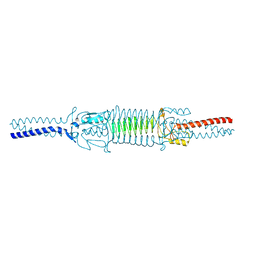 | | Acinetobacter sp. Tol 5 AtaA C-terminal Ylhead fused to GCN4 adaptors (Chead) | | Descriptor: | Trimeric autotransporter adhesin | | Authors: | Koiwai, K, Hartmann, M.D, Yoshimoto, S, Nur 'Izzah, N, Suzuki, A, Linke, D, Lupas, A.N, Hori, K. | | Deposit date: | 2014-01-10 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis for Toughness and Flexibility in the C-terminal Passenger Domain of an Acinetobacter Trimeric Autotransporter Adhesin.
J.Biol.Chem., 291, 2016
|
|
3WPO
 
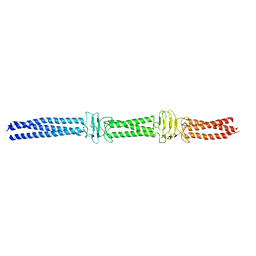 | | Acinetobacter sp. Tol 5 AtaA YDD-DALL3 domains in C-terminal stalk fused to GCN4 adaptors (CstalkC1i) | | Descriptor: | CHLORIDE ION, Trimeric autotransporter adhesin | | Authors: | Koiwai, K, Hartmann, M.D, Yoshimoto, S, Nur 'Izzah, N, Suzuki, A, Linke, D, Lupas, A.N, Hori, K. | | Deposit date: | 2014-01-14 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Structural Basis for Toughness and Flexibility in the C-terminal Passenger Domain of an Acinetobacter Trimeric Autotransporter Adhesin.
J.Biol.Chem., 291, 2016
|
|
3WPA
 
 | | Acinetobacter sp. Tol 5 AtaA C-terminal stalk_FL fused to GCN4 adaptors (CstalkFL) | | Descriptor: | CHLORIDE ION, Trimeric autotransporter adhesin | | Authors: | Koiwai, K, Hartmann, M.D, Yoshimoto, S, Nur 'Izzah, N, Suzuki, A, Linke, D, Lupas, A.N, Hori, K. | | Deposit date: | 2014-01-10 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis for Toughness and Flexibility in the C-terminal Passenger Domain of an Acinetobacter Trimeric Autotransporter Adhesin.
J.Biol.Chem., 291, 2016
|
|
5YV5
 
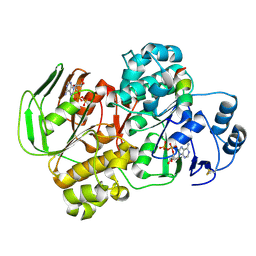 | | Crystal structure of the complex of archaeal ribosomal stalk protein aP1 and archaeal ribosome recycling factor aABCE1. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase RIL, Archaeal ribosomal stalk protein aP1, ... | | Authors: | Imai, H, Abe, T, Miyoshi, T, Nishikawa, S, Ito, K, Uchiumi, T. | | Deposit date: | 2017-11-24 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The ribosomal stalk protein is crucial for the action of the conserved ATPase ABCE1
Nucleic Acids Res., 46, 2018
|
|
1ULW
 
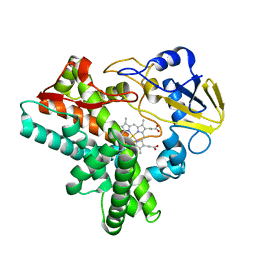 | | Crystal structure of P450nor Ser73Gly/Ser75Gly mutant | | Descriptor: | Cytochrome P450 55A1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Oshima, R, Fushinobu, S, Su, F, Li, Z, Takaya, N, Shoun, H. | | Deposit date: | 2003-09-16 | | Release date: | 2004-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for direct hydride transfer from NADH to cytochrome P450nor
J.Mol.Biol., 342, 2004
|
|
1UMG
 
 | | Crystal structure of fructose-1,6-bisphosphatase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM), 385aa long conserved hypothetical protein, ... | | Authors: | Nishimasu, H, Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2003-09-30 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The first crystal structure of the novel class of fructose-1,6-bisphosphatase present in thermophilic archaea.
Structure, 12, 2004
|
|
5Z1L
 
 | | Cryo-EM structure of Methanoccus maripaludis archaellum | | Descriptor: | Flagellin | | Authors: | Meshcheryakov, V.A, Shibata, S, Schreiber, M.T, Villar-Briones, A, Jarrell, K.F, Aizawa, S, Wolf, M. | | Deposit date: | 2017-12-26 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | High-resolution archaellum structure reveals a conserved metal-binding site.
Embo Rep., 20, 2019
|
|
5X6S
 
 | | Acetyl xylan esterase from Aspergillus awamori | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylxylan esterase A, ... | | Authors: | Komiya, D, Koseki, T, Fushinobu, S. | | Deposit date: | 2017-02-23 | | Release date: | 2017-08-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure and Substrate Specificity Modification of Acetyl Xylan Esterase from Aspergillus luchuensis
Appl. Environ. Microbiol., 83, 2017
|
|
3WQA
 
 | | Acinetobacter sp. Tol 5 AtaA YDD-DALL3 domains in C-terminal stalk fused to GCN4 adaptors (CstalkC1ii) | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, PHOSPHATE ION, ... | | Authors: | Koiwai, K, Hartmann, M.D, Yoshimoto, S, Nur 'Izzah, N, Suzuki, A, Linke, D, Lupas, A.N, Hori, K. | | Deposit date: | 2014-01-24 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural Basis for Toughness and Flexibility in the C-terminal Passenger Domain of an Acinetobacter Trimeric Autotransporter Adhesin.
J.Biol.Chem., 291, 2016
|
|
3WPP
 
 | | Acinetobacter sp. Tol 5 AtaA YDD-DALL3 domains in C-terminal stalk fused to GCN4 adaptors (CstalkC1iii) | | Descriptor: | CHLORIDE ION, Trimeric autotransporter adhesin | | Authors: | Koiwai, K, Hartmann, M.D, Yoshimoto, S, Nur 'Izzah, N, Suzuki, A, Linke, D, Lupas, A.N, Hori, K. | | Deposit date: | 2014-01-15 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Structural Basis for Toughness and Flexibility in the C-terminal Passenger Domain of an Acinetobacter Trimeric Autotransporter Adhesin.
J.Biol.Chem., 291, 2016
|
|
3WPR
 
 | | Acinetobacter sp. Tol 5 AtaA N-terminal half of C-terminal stalk fused to GCN4 adaptors (CstalkN) | | Descriptor: | Trimeric autotransporter adhesin | | Authors: | Koiwai, K, Hartmann, M.D, Yoshimoto, S, Nur 'Izzah, N, Suzuki, A, Linke, D, Lupas, A.N, Hori, K. | | Deposit date: | 2014-01-15 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structural Basis for Toughness and Flexibility in the C-terminal Passenger Domain of an Acinetobacter Trimeric Autotransporter Adhesin.
J.Biol.Chem., 291, 2016
|
|
2QHM
 
 | | crystal structure of Chek1 in complex with inhibitor 2a | | Descriptor: | (3-ENDO)-8-METHYL-8-AZABICYCLO[3.2.1]OCT-3-YL 1H-PYRROLO[2,3-B]PYRIDINE-3-CARBOXYLATE, Serine/threonine-protein kinase Chk1 | | Authors: | Yan, Y, Munshi, S. | | Deposit date: | 2007-07-02 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Optimization of a pyrazoloquinolinone class of Chk1 kinase inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
5YB7
 
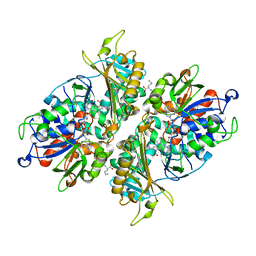 | | L-Amino acid oxidase/monooxygenase from Pseudomonas sp. AIU 813 - L-ornithine complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-amino acid oxidase/monooxygenase, L-ornithine | | Authors: | Im, D, Matsui, D, Arakawa, T, Isobe, K, Asano, Y, Fushinobu, S. | | Deposit date: | 2017-09-03 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand complex structures of l-amino acid oxidase/monooxygenase from
FEBS Open Bio, 8, 2018
|
|
5YHS
 
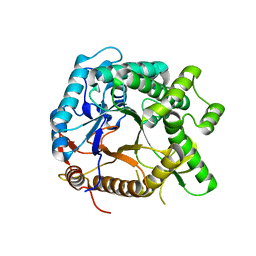 | | Pyruvylated beta-D-galactosidase from Bacillus sp. HMA207, apo form | | Descriptor: | Pyruvylated beta-D-galactosidase | | Authors: | Tanuma, M, Yamada, C, Arakawa, T, Higuchi, Y, Takegawa, K, Fushinobu, S. | | Deposit date: | 2017-09-29 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification and characterization of a novel beta-D-galactosidase that releases pyruvylated galactose.
Sci Rep, 8, 2018
|
|
5YIF
 
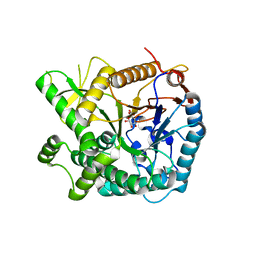 | | Pyruvylated beta-D-galactosidase from Bacillus sp. HMA207, E163A mutant pyruvylated beta-D-galactose complex | | Descriptor: | (2R,4aR,6R,7R,8R,8aR)-2-methyl-6,7,8-tris(oxidanyl)-4,4a,6,7,8,8a-hexahydropyrano[3,2-d][1,3]dioxine-2-carboxylic acid, Pyruvylated beta-D-galactosidase | | Authors: | Tanuma, M, Yamada, C, Arakawa, T, Higuchi, Y, Takegawa, K, Fushinobu, S. | | Deposit date: | 2017-10-04 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Identification and characterization of a novel beta-D-galactosidase that releases pyruvylated galactose.
Sci Rep, 8, 2018
|
|
5YSD
 
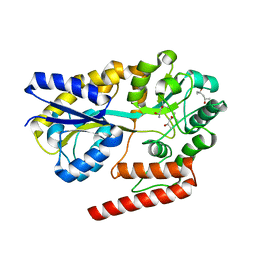 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophorotriose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5YB6
 
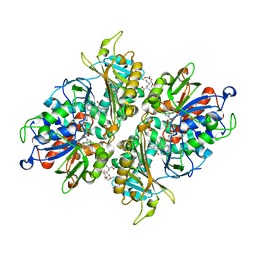 | | L-Amino acid oxidase/monooxygenase from Pseudomonas sp. AIU 813 - L-lysine complex | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, FLAVIN-ADENINE DINUCLEOTIDE, L-amino acid oxidase/monooxygenase, ... | | Authors: | Im, D, Matsui, D, Arakawa, T, Isobe, K, Asano, Y, Fushinobu, S. | | Deposit date: | 2017-09-03 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand complex structures of l-amino acid oxidase/monooxygenase from
FEBS Open Bio, 8, 2018
|
|
5YSF
 
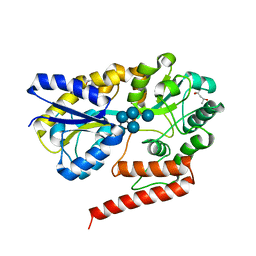 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophoropentaose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5YB8
 
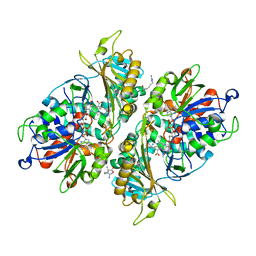 | | L-Amino acid oxidase/monooxygenase from Pseudomonas sp. AIU 813 - L-arginine complex | | Descriptor: | ARGININE, FLAVIN-ADENINE DINUCLEOTIDE, L-amino acid oxidase/monooxygenase | | Authors: | Im, D, Matsui, D, Arakawa, T, Isobe, K, Asano, Y, Fushinobu, S. | | Deposit date: | 2017-09-03 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ligand complex structures of l-amino acid oxidase/monooxygenase from
FEBS Open Bio, 8, 2018
|
|
5YSE
 
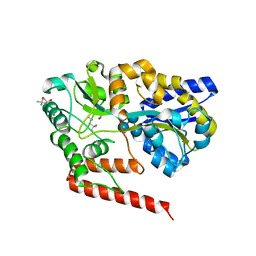 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophorotetraose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5YSB
 
 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in ligand-free form | | Descriptor: | DI(HYDROXYETHYL)ETHER, Lin1841 protein, ZINC ION | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-13 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5WL0
 
 | | Co-crystal structure of Influenza A H3N2 PB2 (241-741) bound to VX-787 | | Descriptor: | (2S,3S)-3-[[5-fluoranyl-2-(5-fluoranyl-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-4-yl]amino]bicyclo[2.2.2]octane-2-carboxylic acid, DI(HYDROXYETHYL)ETHER, Polymerase basic protein 2 | | Authors: | Ma, X, Shia, S. | | Deposit date: | 2017-07-25 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for therapeutic inhibition of influenza A polymerase PB2 subunit.
Sci Rep, 7, 2017
|
|
1V5C
 
 | | The crystal structure of the inactive form chitosanase from Bacillus sp. K17 at pH3.7 | | Descriptor: | SULFATE ION, chitosanase | | Authors: | Adachi, W, Shimizu, S, Sunami, T, Fukazawa, T, Suzuki, M, Yatsunami, R, Nakamura, S, Takenaka, A. | | Deposit date: | 2003-11-22 | | Release date: | 2004-12-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of family GH-8 chitosanase with subclass II specificity from Bacillus sp. K17
J.MOL.BIOL., 343, 2004
|
|
5Z8T
 
 | | Flavin-containing monooxygenase | | Descriptor: | Acyl-CoA dehydrogenase type 2 domain protein, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Choe, J, Moon, H, Shin, S. | | Deposit date: | 2018-02-01 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of putative Flavin-dependent monooxgenase
To Be Published
|
|
