6UR1
 
 | |
6UQZ
 
 | |
6UQY
 
 | |
6UR0
 
 | | Crystal structure of ChoE D285N mutant acyl-enzyme | | Descriptor: | 2-(TRIMETHYLAMMONIUM)ETHYL THIOL, ChoE, GLYCEROL | | Authors: | Pham, V.D, Shi, R. | | Deposit date: | 2019-10-21 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the putative bacterial acetylcholinesterase ChoE and its substrate inhibition mechanism.
J.Biol.Chem., 295, 2020
|
|
3O39
 
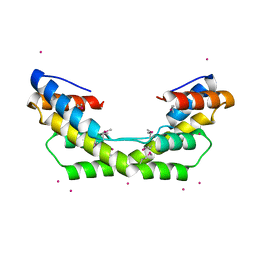 | |
4X8Q
 
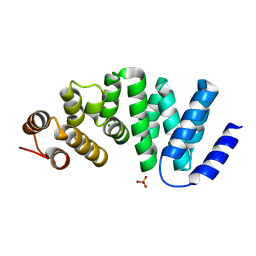 | | X-ray crystal structure of AlkD2 from Streptococcus mutans | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Mullins, E.A, Shi, R, Eichman, B.F. | | Deposit date: | 2014-12-10 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | A New Family of HEAT-Like Repeat Proteins Lacking a Critical Substrate Recognition Motif Present in Related DNA Glycosylases.
Plos One, 10, 2015
|
|
3OQ5
 
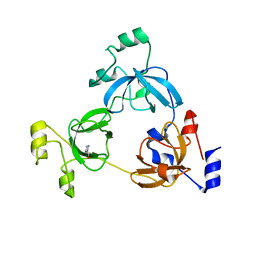 | | Crystal structure of the 3-MBT domain from human L3MBTL1 in complex with p53K382me1 | | Descriptor: | Cellular tumor antigen p53, Lethal(3)malignant brain tumor-like protein | | Authors: | Roy, S, West, L.E, Weiner, K.L, Hayashi, R, Shi, X, Appella, E, Gozani, O, Kutateladze, T. | | Deposit date: | 2010-09-02 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5005 Å) | | Cite: | The MBT Repeats of L3MBTL1 Link SET8-mediated p53 Methylation at Lysine 382 to Target Gene Repression.
J.Biol.Chem., 285, 2010
|
|
5JMF
 
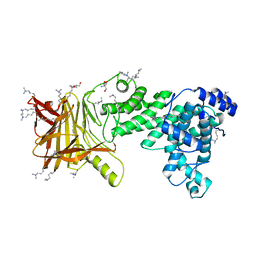 | | Heparinase III-BT4657 gene product | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Heparinase III protein, ... | | Authors: | Ulaganathan, T.S, Shi, R, Yao, D, Garron, M.-L, Cherney, M, Cygler, M. | | Deposit date: | 2016-04-28 | | Release date: | 2016-05-25 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | Conformational flexibility of PL12 family heparinases: structure and substrate specificity of heparinase III from Bacteroides thetaiotaomicron (BT4657).
Glycobiology, 27, 2017
|
|
5JMD
 
 | | Heparinase III-BT4657 gene product, Methylated Lysines | | Descriptor: | Heparinase III protein, MAGNESIUM ION | | Authors: | Ulaganathan, T.S, Shi, R, Yao, D, Garron, M.-L, Cherney, M, Cygler, M. | | Deposit date: | 2016-04-28 | | Release date: | 2016-05-25 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational flexibility of PL12 family heparinases: structure and substrate specificity of heparinase III from Bacteroides thetaiotaomicron (BT4657).
Glycobiology, 27, 2017
|
|
5I1W
 
 | | Crystal structure of CrmK, a flavoenzyme involved in the shunt product recycling mechanism in caerulomycin biosynthesis | | Descriptor: | 4-hydroxy[2,2'-bipyridine]-6-carbaldehyde, 6-(hydroxymethyl)[2,2'-bipyridin]-4-ol, CrmK, ... | | Authors: | Picard, M.-E, Barma, J, Shi, R. | | Deposit date: | 2016-02-07 | | Release date: | 2017-02-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Biochemical and structural insights into flavoenzyme CrmK reveals a shunt product recycling mechanism in caerulomycin biosynthesis
to be published
|
|
8VLM
 
 | | Crystal structure of the yeast cytosine deaminase (yCD) E64V-M100W heterodimer | | Descriptor: | 1,2-ETHANEDIOL, Cytosine deaminase, ZINC ION | | Authors: | Picard, M.-E, Grenier, G, Despres, P.C, Dube, A.K, Landry, C.R, Shi, R. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Compensatory mutations potentiate constructive neutral evolution by gene duplication.
Science, 385, 2024
|
|
8VLK
 
 | | Crystal structure of the yeast cytosine deaminase containing both open and closed active sites | | Descriptor: | 1,2-ETHANEDIOL, Cytosine deaminase, SULFATE ION, ... | | Authors: | Picard, M.-E, Grenier, J, Despres, P.C, Dube, A.K, Landry, C.R, Shi, R. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Compensatory mutations potentiate constructive neutral evolution by gene duplication.
Science, 385, 2024
|
|
8VLJ
 
 | | Crystal structure of the cacodylate-bound yeast cytosine deaminase (closed form) | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, Cytosine deaminase, ... | | Authors: | Picard, M.-E, Grenier, J, Despres, P.C, Dube, A.K, Landry, C.R, Shi, R. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Compensatory mutations potentiate constructive neutral evolution by gene duplication.
Science, 385, 2024
|
|
8VLL
 
 | | Crystal structure of the yeast cytosine deaminase (yCD) M100W mutant | | Descriptor: | 1,2-ETHANEDIOL, Cytosine deaminase, PHOSPHATE ION, ... | | Authors: | Picard, M.-E, Grenier, J, Despres, P.C, Dube, A.K, Landry, C.R, Shi, R. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Compensatory mutations potentiate constructive neutral evolution by gene duplication.
Science, 385, 2024
|
|
6M9M
 
 | |
5I1V
 
 | |
6B02
 
 | |
6B07
 
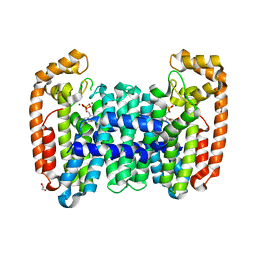 | | Crystal structure of CfFPPS2, a lepidopteran type-II farnesyl diphosphate synthase, complexed with [1-phosphono-2-(1-propylpyridin-2-yl)ethyl]phosphonic acid (inhibitor 1d) | | Descriptor: | 1,2-ETHANEDIOL, 2-(2,2-diphosphonoethyl)-1-propylpyridin-1-ium, Farnesyl diphosphate synthase, ... | | Authors: | Picard, M.-E, Cusson, M, Shi, R. | | Deposit date: | 2017-09-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural characterization of a lepidopteran type-II farnesyl diphosphate synthase from the spruce budworm, Choristoneura fumiferana: Implications for inhibitor design.
Insect Biochem. Mol. Biol., 92, 2017
|
|
6B06
 
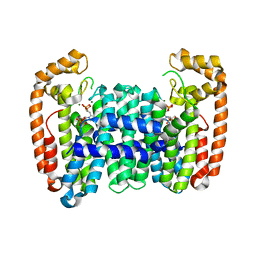 | | Crystal structure of CfFPPS2, a lepidopteran type-II farnesyl diphosphate synthase, complexed with IPP and [2-(1-methylpyridin-2-yl)-1-phosphono-ethyl]phosphonic acid (inhibitor 1b) | | Descriptor: | 2-(2,2-diphosphonoethyl)-1-methylpyridin-1-ium, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Farnesyl diphosphate synthase, ... | | Authors: | Picard, M.-E, Cusson, M, Shi, R. | | Deposit date: | 2017-09-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural characterization of a lepidopteran type-II farnesyl diphosphate synthase from the spruce budworm, Choristoneura fumiferana: Implications for inhibitor design.
Insect Biochem. Mol. Biol., 92, 2017
|
|
6B04
 
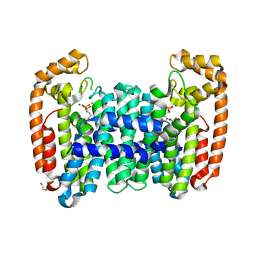 | | Crystal structure of CfFPPS2, a lepidopteran type-II farnesyl diphosphate synthase, complexed with [2-(1-methylpyridin-2-yl)-1-phosphono-ethyl]phosphonic acid (inhibitor 1b) | | Descriptor: | 1,2-ETHANEDIOL, 2-(2,2-diphosphonoethyl)-1-methylpyridin-1-ium, Farnesyl diphosphate synthase, ... | | Authors: | Picard, M.-E, Cusson, M, Shi, R. | | Deposit date: | 2017-09-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural characterization of a lepidopteran type-II farnesyl diphosphate synthase from the spruce budworm, Choristoneura fumiferana: Implications for inhibitor design.
Insect Biochem. Mol. Biol., 92, 2017
|
|
7VZT
 
 | | A human neutralizing antibody targeting SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GH12-Heavy, GH12-LIGHT, ... | | Authors: | Wang, F.Z, Wang, Y, Tan, X.W, Shi, R, Yan, J.H. | | Deposit date: | 2021-11-16 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | GH12, glycosylation function
To Be Published
|
|
1OV4
 
 | | Crystal structure of human DHEA-ST complexed with androsterone | | Descriptor: | (3Beta,5alpha)-3-Hydroxyandrostan-17-one, Alcohol sulfotransferase, SULFATE ION | | Authors: | Chang, H.J, Shi, R, Rhese, P, Lin, S.X. | | Deposit date: | 2003-03-25 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identifying androsterone (ADT) as a cognate substrate for human dehydroepiandrosterone sulfotransferase (DHEA-ST) important for steroid homeostasis: structure of the enzyme-ADT complex.
J.Biol.Chem., 279, 2004
|
|
7C88
 
 | | Complex structure of JS003 and PD-L1 | | Descriptor: | JS003 Heavy chain, JS003 Light chain, Programmed cell death 1 ligand 1 | | Authors: | Bi, X, Shi, R, Chai, Y, Qi, J, Yan, J, Tan, S. | | Deposit date: | 2020-05-29 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Identification of a hotspot on PD-L1 for pH-dependent binding by monoclonal antibodies for tumor therapy.
Signal Transduct Target Ther, 5, 2020
|
|
4X5M
 
 | | Crystal structure of SemiSWEET in the inward-open conformation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, OLEIC ACID, ... | | Authors: | Lee, Y, Nishizawa, T, Yamashita, K, Ishitani, R, Nureki, O. | | Deposit date: | 2014-12-05 | | Release date: | 2015-01-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the facilitative diffusion mechanism by SemiSWEET transporter
Nat Commun, 6, 2015
|
|
4URL
 
 | | Crystal Structure of Staph ParE43kDa in complex with KBD | | Descriptor: | (1R,4aS,5S,6S,8aR)-5-{[(5S)-1-(3-O-acetyl-4-O-carbamoyl-6-deoxy-2-O-methyl-alpha-L-talopyranosyl)-4-hydroxy-2-oxo-5-(propan-2-yl)-2,5-dihydro-1H-pyrrol-3-yl]carbonyl}-6-methyl-4-methylidene-1,2,3,4,4a,5,6,8a-octahydronaphthalen-1-yl 2,6-dideoxy-3-C-[(1S)-1-{[(3,4-dichloro-5-methyl-1H-pyrrol-2-yl)carbonyl]amino}ethyl]-beta-D-ribo-hexopyranoside, DNA TOPOISOMERASE IV, B SUBUNIT | | Authors: | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | Deposit date: | 2014-06-30 | | Release date: | 2014-07-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
