4U4T
 
 | | Structure of a nitrate/nitrite antiporter NarK in nitrate-bound inward-open state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, NITRATE ION, Nitrate/nitrite transporter NarK, ... | | Authors: | Fukuda, M, Takeda, H, Kato, H.E, Doki, S, Ito, K, Maturana, A.D, Ishitani, R, Nureki, O. | | Deposit date: | 2014-07-24 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for dynamic mechanism of nitrate/nitrite antiport by NarK
Nat Commun, 6, 2015
|
|
4UDD
 
 | | GR in complex with desisobutyrylciclesonide | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, DESISOBUYTYRYL CICLESONIDE, ... | | Authors: | Edman, K, Hogner, A, Hussein, A, Bjursell, M, Aagaard, A, Backstrom, S, Bodin, C, Wissler, L, Jellesmark-Jensen, T, Cavallin, A, Karlsson, U, Nilsson, E, Lecina, D, Takahashi, R, Grebner, C, Lepisto, M, Guallar, V. | | Deposit date: | 2014-12-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ligand Binding Mechanism in Steroid Receptors: From Conserved Plasticity to Differential Evolutionary Constraints.
Structure, 23, 2015
|
|
4UDC
 
 | | GR in complex with dexamethasone | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, DEXAMETHASONE, GLUCOCORTICOID RECEPTOR, ... | | Authors: | Edman, K, Hogner, A, Hussein, A, Bjursell, M, Aagaard, A, Backstrom, S, Bodin, C, Wissler, L, Jellesmark-Jensen, T, Cavallin, A, Karlsson, U, Nilsson, E, Lecina, D, Takahashi, R, Grebner, C, Lepisto, M, Guallar, V. | | Deposit date: | 2014-12-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand Binding Mechanism in Steroid Receptors: From Conserved Plasticity to Differential Evolutionary Constraints.
Structure, 23, 2015
|
|
4UDA
 
 | | MR in complex with dexamethasone | | Descriptor: | DEXAMETHASONE, GLYCEROL, MINERALOCORTICOID RECEPTOR, ... | | Authors: | Edman, K, Hogner, A, Hussein, A, Bjursell, M, Aagaard, A, Backstrom, S, Bodin, C, Wissler, L, Jellesmark-Jensen, T, Cavallin, A, Karlsson, U, Nilsson, E, Lecina, D, Takahashi, R, Grebner, C, Lepisto, M, Guallar, V. | | Deposit date: | 2014-12-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Ligand Binding Mechanism in Steroid Receptors: From Conserved Plasticity to Differential Evolutionary Constraints.
Structure, 23, 2015
|
|
6IU4
 
 | | Crystal structure of iron transporter VIT1 with cobalt ion | | Descriptor: | COBALT (II) ION, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Taniguchi, R, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
6IU9
 
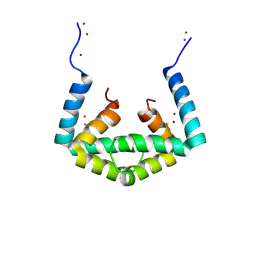 | | Crystal structure of cytoplasmic metal binding domain with iron ions | | Descriptor: | FE (II) ION, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
6IU8
 
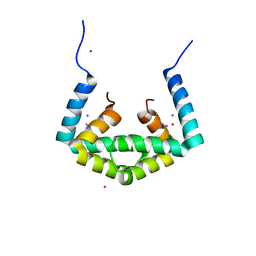 | | Crystal structure of cytoplasmic metal binding domain with cobalt ions | | Descriptor: | COBALT (II) ION, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
6IU6
 
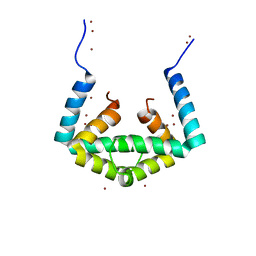 | | Crystal structure of cytoplasmic metal binding domain with nickel ions | | Descriptor: | NICKEL (II) ION, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
1B90
 
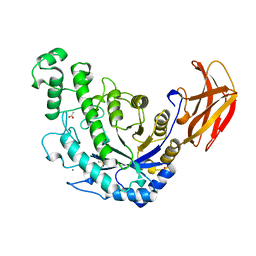 | | BACILLUS CEREUS BETA-AMYLASE APO FORM | | Descriptor: | ACETATE ION, CALCIUM ION, PROTEIN (BETA-AMYLASE), ... | | Authors: | Mikami, B, Adachi, M, Kage, T, Sarikaya, E, Nanmori, T, Shinke, R, Utsumi, S. | | Deposit date: | 1999-03-06 | | Release date: | 1999-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of raw starch-digesting Bacillus cereus beta-amylase complexed with maltose.
Biochemistry, 38, 1999
|
|
8K5W
 
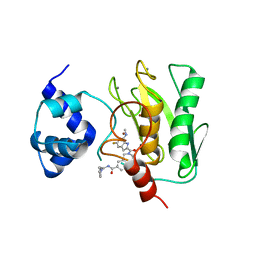 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | Descriptor: | 2-[[5-fluoranyl-7-(methylamino)-1H-indol-2-yl]carbonyl]-N-(2-pyrrol-1-ylethyl)-3,4-dihydro-1H-isoquinoline-7-carboxamide, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
8K5V
 
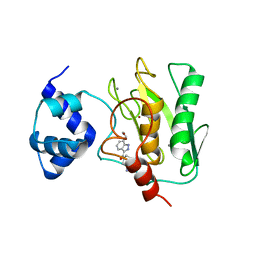 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | Descriptor: | 6,7-dihydro-4H-[1,3]oxazolo[4,5-c]pyridin-5-yl-(7-ethyl-2H-indazol-3-yl)methanone, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
8K5Y
 
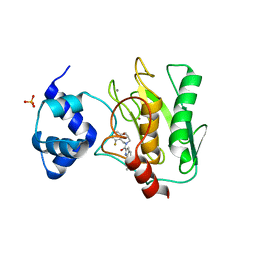 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | Descriptor: | (3-azanyl-4-fluoranyl-5,7,8,9-tetrahydropyrido[4,3-c]azepin-6-yl)-[6-(2-oxidanylpropan-2-yl)-1H-indol-2-yl]methanone, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
8K5X
 
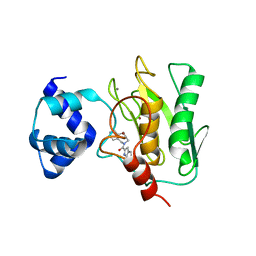 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | Descriptor: | (6-cyclopropyl-1~{H}-indol-2-yl)-(5,7,8,9-tetrahydropyrido[4,3-c]azepin-6-yl)methanone, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
1B9Z
 
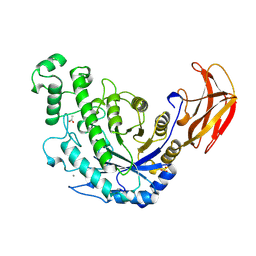 | | BACILLUS CEREUS BETA-AMYLASE COMPLEXED WITH MALTOSE | | Descriptor: | ACETATE ION, CALCIUM ION, PROTEIN (BETA-AMYLASE), ... | | Authors: | Mikami, B, Adachi, M, Kage, T, Sarikaya, E, Nanmori, T, Shinke, R, Utsumi, S. | | Deposit date: | 1999-03-06 | | Release date: | 1999-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of raw starch-digesting Bacillus cereus beta-amylase complexed with maltose.
Biochemistry, 38, 1999
|
|
6JMR
 
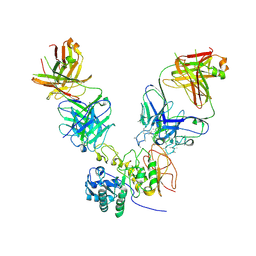 | | CD98hc extracellular domain bound to HBJ127 Fab and MEM-108 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | Authors: | Lee, Y, Nishizawa, T, Kusakizako, T, Oda, K, Ishitani, R, Yokoyama, T, Nakane, T, Shirouzu, M, Nureki, O. | | Deposit date: | 2019-03-13 | | Release date: | 2019-06-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the human L-type amino acid transporter 1 in complex with glycoprotein CD98hc.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6IU5
 
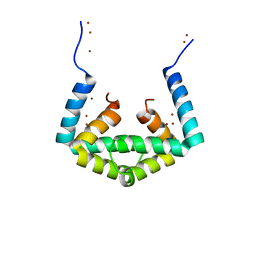 | | Crystal structure of cytoplasmic metal binding domain with zinc ions | | Descriptor: | CHLORIDE ION, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
5XQR
 
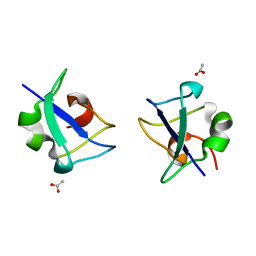 | | Crystal structure of Notched-fin eelpout type III antifreeze protein A20V mutant (NFE6, AFP), C2221 form | | Descriptor: | ACETATE ION, Ice-structuring protein | | Authors: | Adachi, M, Shimizu, R, Shibazaki, C, Kondo, H, Tsuda, S. | | Deposit date: | 2017-06-07 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Polypentagonal ice-like water networks emerge solely in an activity-improved variant of ice-binding protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5XQU
 
 | | Crystal structure of Notched-fin eelpout type III antifreeze protein A20I mutant (NFE6, AFP), P212121 form | | Descriptor: | Ice-structuring protein | | Authors: | Adachi, M, Shimizu, R, Shibazaki, C, Kondo, H, Tsuda, S. | | Deposit date: | 2017-06-07 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Polypentagonal ice-like water networks emerge solely in an activity-improved variant of ice-binding protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5XQV
 
 | | Crystal structure of Notched-fin eelpout type III antifreeze protein A20L mutant (NFE6, AFP), P21 form | | Descriptor: | Ice-structuring protein | | Authors: | Adachi, M, Shimizu, R, Shibazaki, C, Kondo, H, Tsuda, S. | | Deposit date: | 2017-06-07 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Polypentagonal ice-like water networks emerge solely in an activity-improved variant of ice-binding protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5XR0
 
 | | Crystal structure of Notched-fin eelpout type III antifreeze protein A20T mutant (NFE6, AFP), P21 form | | Descriptor: | Ice-structuring protein | | Authors: | Adachi, M, Shimizu, R, Shibazaki, C, Kondo, H, Tsuda, S. | | Deposit date: | 2017-06-07 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Polypentagonal ice-like water networks emerge solely in an activity-improved variant of ice-binding protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5XQP
 
 | | Crystal structure of Notched-fin eelpout type III antifreeze protein (NFE6, AFP), P212121 form | | Descriptor: | Ice-structuring protein, SULFATE ION | | Authors: | Adachi, M, Shimizu, R, Shibazaki, C, Kondo, H, Tsuda, S. | | Deposit date: | 2017-06-07 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Polypentagonal ice-like water networks emerge solely in an activity-improved variant of ice-binding protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7RZN
 
 | | Crystal Structure of Ferritin grown by microbatch method in presence of agarose and electric field 2.1KV | | Descriptor: | CADMIUM ION, Ferritin light chain, SULFATE ION | | Authors: | Aditya, S, Priyadharshine, R, Maham, I, Miller, J.D, Stojanoff, V. | | Deposit date: | 2021-08-27 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of Ferritin grown by microbatch method in presence of agarose and electric field 2.1KV
To Be Published
|
|
7RYW
 
 | | Crystal structure of Ferritin grown by the microbatch method in the presence of Agarose | | Descriptor: | CADMIUM ION, Ferritin light chain, SULFATE ION | | Authors: | Aditya, S, Priyadharshine, R, Maham, I, Miller, D.J, Stojanoff, V. | | Deposit date: | 2021-08-26 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of Ferritin grown by microbatch method in the presence of agarose
To Be Published
|
|
7RZX
 
 | | Crystal Structure of Ferritin grown by microbatch method in presence of agarose and electric field 4.3KV | | Descriptor: | CADMIUM ION, Ferritin light chain, SULFATE ION | | Authors: | Aditya, S, Priyadharshine, R, Maham, I, Miller, J.D, Stojanoff, V. | | Deposit date: | 2021-08-27 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal Structure of Ferritin grown by microbatch method in presence of agarose and electric field 4.3KV
To Be Published
|
|
3WS4
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-2A) | | Descriptor: | Beta-lactamase, CHLORIDE ION, STRONTIUM ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-28 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
