8IAW
 
 | |
8IAU
 
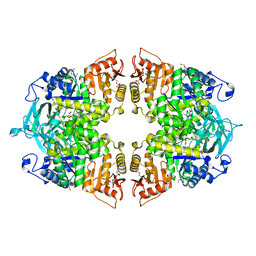 | | Crystal structure of Streptococcus pneumoniae pyruvate kinase in complex with oxalate and fructose 1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Nakashima, R, Taguchi, A. | | Deposit date: | 2023-02-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of Streptococcus pneumoniae pyruvate kinase involved in fosfomycin resistance.
J.Biol.Chem., 299, 2023
|
|
8IAV
 
 | |
6IE9
 
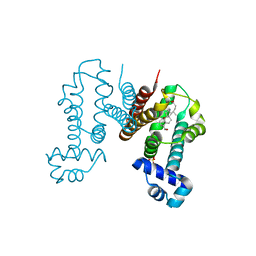 | | RamR in complex with chenodeoxycholic acid | | Descriptor: | CHENODEOXYCHOLIC ACID, Regulatory protein, SULFATE ION | | Authors: | Nakashima, R, Sakurai, K, Yamasaki, S, Nishino, K. | | Deposit date: | 2018-09-13 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of the multidrug resistance regulator RamR complexed with bile acids.
Sci Rep, 9, 2019
|
|
2E5D
 
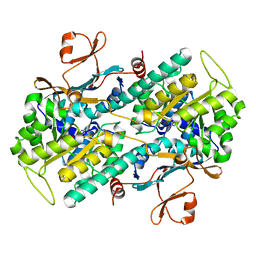 | | Crystal structure of Human NMPRTase complexed with nicotinamide | | Descriptor: | NICOTINAMIDE, Nicotinamide phosphoribosyltransferase | | Authors: | Takahashi, R, Nakamura, S, Kobayashi, Y, Ohkubo, T. | | Deposit date: | 2006-12-20 | | Release date: | 2007-12-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and reaction mechanism of human nicotinamide phosphoribosyltransferase
J.Biochem., 147, 2010
|
|
2E5C
 
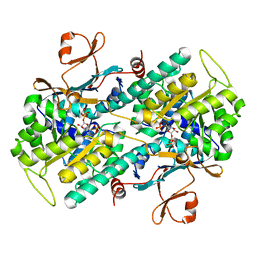 | | Crystal structure of Human NMPRTase complexed with 5'-phosphoribosyl-1'-pyrophosphate | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, Nicotinamide phosphoribosyltransferase | | Authors: | Takahashi, R, Nakamura, S, Kobayashi, Y, Ohkubo, T. | | Deposit date: | 2006-12-20 | | Release date: | 2007-12-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and reaction mechanism of human nicotinamide phosphoribosyltransferase
J.Biochem., 147, 2010
|
|
7D5R
 
 | | Structure of the Ca2+-bound C646A mutant of peptidylarginine deiminase type III (PAD3) | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Mashimo, R, Akimoto, M, Unno, M. | | Deposit date: | 2020-09-28 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.148 Å) | | Cite: | Structures of human peptidylarginine deiminase type III provide insights into substrate recognition and inhibitor design.
Arch.Biochem.Biophys., 708, 2021
|
|
3OOL
 
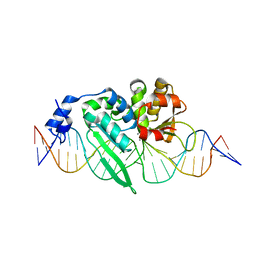 | | I-SceI complexed with C/G+4 DNA substrate | | Descriptor: | 5'-D(*CP*AP*CP*GP*CP*TP*AP*GP*GP*GP*AP*TP*AP*AP*CP*CP*GP*GP*GP*TP*AP*AP*TP*AP*C)-3', 5'-D(*GP*GP*TP*AP*TP*TP*AP*CP*CP*CP*GP*GP*TP*TP*AP*TP*CP*CP*CP*TP*AP*GP*CP*GP*T)-3', CALCIUM ION, ... | | Authors: | Joshi, R, Chen, J.-H, Golden, B.L, Gimble, F.S. | | Deposit date: | 2010-08-31 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of I-SceI Homing Endonucleases with Increased DNA Recognition Site Specificity.
J.Mol.Biol., 405, 2011
|
|
7CAO
 
 | | Crystal structure of red chromoprotein from Olindias formosa | | Descriptor: | Chromoprotein | | Authors: | Nakashima, R, Zhai, L, Ike, Y, Matsudz, T, Nagai, T. | | Deposit date: | 2020-06-09 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-based analysis and evolution of a monomerized red-colored chromoprotein from the Olindias formosa jellyfish.
Protein Sci., 31, 2022
|
|
3OOR
 
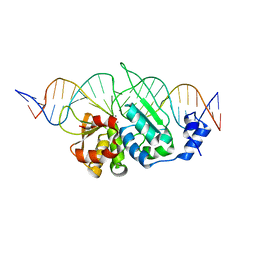 | | I-SceI mutant (K86R/G100T)complexed with C/G+4 DNA substrate | | Descriptor: | 5'-D(*CP*AP*CP*GP*CP*TP*AP*GP*GP*GP*AP*TP*AP*AP*CP*CP*GP*GP*GP*TP*AP*AP*TP*AP*C)-3', 5'-D(*GP*GP*TP*AP*TP*TP*AP*CP*CP*CP*GP*GP*TP*TP*AP*TP*CP*CP*CP*TP*AP*GP*CP*GP*T)-3', CALCIUM ION, ... | | Authors: | Joshi, R, Chen, J.-H, Golden, B.L, Gimble, F.S. | | Deposit date: | 2010-08-31 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evolution of I-SceI Homing Endonucleases with Increased DNA Recognition Site Specificity.
J.Mol.Biol., 405, 2011
|
|
2EY4
 
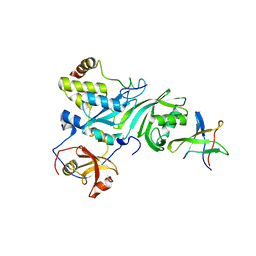 | | Crystal Structure of a Cbf5-Nop10-Gar1 Complex | | Descriptor: | Probable tRNA pseudouridine synthase B, Ribosome biogenesis protein Nop10, ZINC ION, ... | | Authors: | Rashid, R, Liang, B, Li, H, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-11-09 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of a Cbf5-Nop10-Gar1 complex and implications in RNA-guided pseudouridylation and dyskeratosis congenita.
Mol.Cell, 21, 2006
|
|
6IIA
 
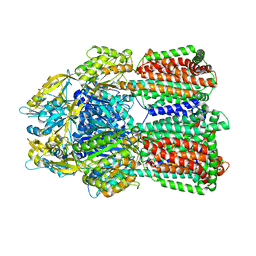 | | MexB in complex with LMNG | | Descriptor: | Lauryl Maltose Neopentyl Glycol, Multidrug resistance protein MexB | | Authors: | Nakashima, R, Sakurai, K, Nakao, K. | | Deposit date: | 2018-10-04 | | Release date: | 2019-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structures of multidrug efflux pump MexB bound with high-molecular-mass compounds.
Sci Rep, 9, 2019
|
|
1Q9P
 
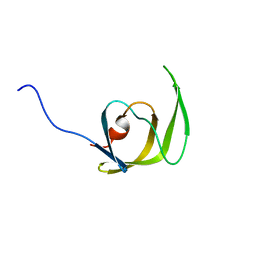 | | Solution structure of the mature HIV-1 protease monomer | | Descriptor: | HIV-1 Protease | | Authors: | Ishima, R, Torchia, D.A, Lynch, S.M, Gronenborn, A.M, Louis, J.M. | | Deposit date: | 2003-08-25 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the mature HIV-1 protease monomer: Insight into the tertiary fold and stability of a precursor
J.Biol.Chem., 278, 2003
|
|
1UDS
 
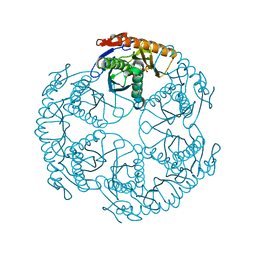 | | Crystal structure of the tRNA processing enzyme RNase PH R126A mutant from Aquifex aeolicus | | Descriptor: | PHOSPHATE ION, Ribonuclease PH, SULFATE ION | | Authors: | Ishii, R, Nureki, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-02 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the tRNA Processing Enzyme RNase PH from Aquifex aeolicus
J.Biol.Chem., 278, 2003
|
|
1UDN
 
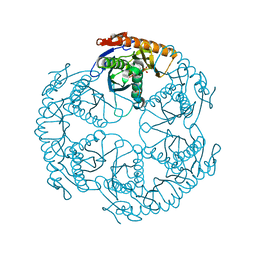 | | Crystal structure of the tRNA processing enzyme RNase PH from Aquifex aeolicus | | Descriptor: | PHOSPHATE ION, Ribonuclease PH, SULFATE ION | | Authors: | Ishii, R, Nureki, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-02 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the tRNA Processing Enzyme RNase PH from Aquifex aeolicus
J.Biol.Chem., 278, 2003
|
|
1UDO
 
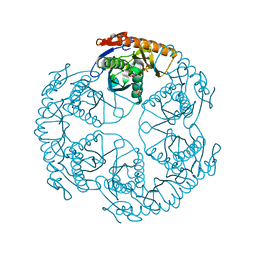 | | Crystal structure of the tRNA processing enzyme RNase PH R86A mutant from Aquifex aeolicus | | Descriptor: | PHOSPHATE ION, Ribonuclease PH, SULFATE ION | | Authors: | Ishii, R, Nureki, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-02 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the tRNA Processing Enzyme RNase PH from Aquifex aeolicus
J.Biol.Chem., 278, 2003
|
|
1UDQ
 
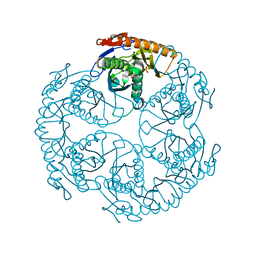 | | Crystal structure of the tRNA processing enzyme RNase PH T125A mutant from Aquifex aeolicus | | Descriptor: | PHOSPHATE ION, Ribonuclease PH, SULFATE ION | | Authors: | Ishii, R, Nureki, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-02 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the tRNA Processing Enzyme RNase PH from Aquifex aeolicus
J.Biol.Chem., 278, 2003
|
|
1WD5
 
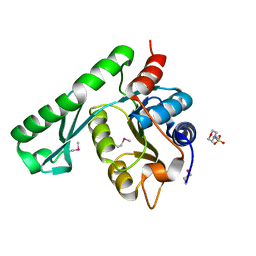 | | Crystal structure of TT1426 from Thermus thermophilus HB8 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, hypothetical protein TT1426 | | Authors: | Shibata, R, Kukimoto-Niino, M, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-11 | | Release date: | 2004-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a predicted phosphoribosyltransferase (TT1426) from Thermus thermophilus HB8 at 2.01 A resolution
Protein Sci., 14, 2005
|
|
2LAB
 
 | |
3W1W
 
 | | Protein-drug complex | | Descriptor: | 1,2-ETHANEDIOL, 2-HYDROXYBENZOIC ACID, CHOLIC ACID, ... | | Authors: | Ishii, R, Gupta, V, Yamaguchi, Y, Handa, H, Nureki, O. | | Deposit date: | 2012-11-21 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Salicylic Acid induces mitochondrial injury by inhibiting ferrochelatase heme biosynthesis activity
Mol.Pharmacol., 84, 2013
|
|
3VYW
 
 | | Crystal structure of MNMC2 from Aquifex Aeolicus | | Descriptor: | BENZAMIDINE, MNMC2, S-ADENOSYLMETHIONINE | | Authors: | Shibata, R, Bessho, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-10-03 | | Release date: | 2012-10-17 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Characterization and structure of the Aquifex aeolicus protein DUF752: a bacterial tRNA-methyltransferase (MnmC2) functioning without the usually fused oxidase domain (MnmC1).
J.Biol.Chem., 287, 2012
|
|
2E7Y
 
 | | High resolution structure of T. maritima tRNase Z | | Descriptor: | S-1,2-PROPANEDIOL, SULFATE ION, ZINC ION, ... | | Authors: | Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-15 | | Release date: | 2007-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The structure of the flexible arm of Thermotoga maritima tRNase Z differs from those of homologous enzymes
Acta Crystallogr.,Sect.F, 63, 2007
|
|
5Y01
 
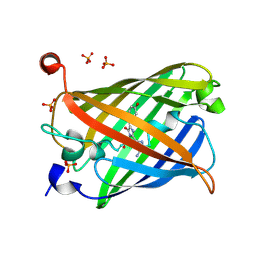 | | Acid-tolerant monomeric GFP, Gamillus, non-fluorescence (OFF) state | | Descriptor: | Green fluorescent protein, PHOSPHATE ION | | Authors: | Nakashima, R, Sakurai, K, Shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2017-07-14 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Acid-Tolerant Monomeric GFP from Olindias formosa.
Cell Chem Biol, 25, 2018
|
|
5Y00
 
 | | Acid-tolerant monomeric GFP, Gamillus, fluorescence (ON) state | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Nakashima, R, Sakurai, K, Shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2017-07-14 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Acid-Tolerant Monomeric GFP from Olindias formosa.
Cell Chem Biol, 25, 2018
|
|
2CY1
 
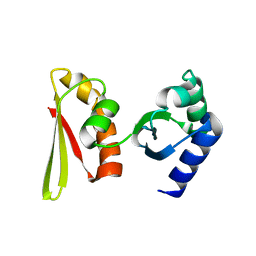 | | Crystal structure of APE1850 | | Descriptor: | NusA protein homolog | | Authors: | Shibata, R, Bessho, Y, Umehara, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallization of the archaeal transcription termination factor NusA: a significant decrease in twinning under microgravity conditions
Acta Crystallogr.,Sect.F, 63, 2007
|
|
