5II4
 
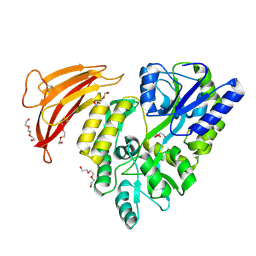 | | Crystal structure of red abalone VERL repeat 1 with linker at 2.0 A resolution | | Descriptor: | Maltose-binding periplasmic protein,Vitelline envelope sperm lysin receptor, TRIETHYLENE GLYCOL, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Sadat Al-Hosseini, H, Raj, I, Nishimura, K, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
5H6O
 
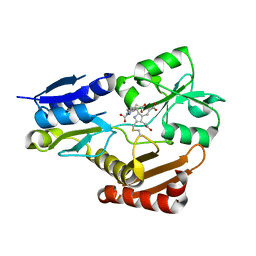 | | Porphobilinogen deaminase from Vibrio Cholerae | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, MAGNESIUM ION, Porphobilinogen deaminase | | Authors: | Funamizu, T, Chen, M, Tanaka, Y, Ishimori, K, Uchida, T. | | Deposit date: | 2016-11-14 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Porphobilinogen deaminase from Vibrio Cholerae
To Be Published
|
|
8IYR
 
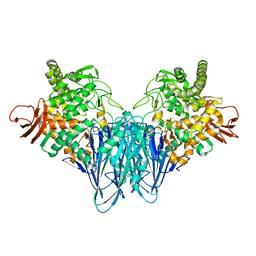 | |
5II6
 
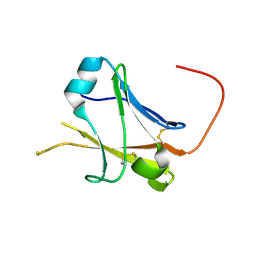 | | Crystal structure of the ZP-N1 domain of mouse sperm receptor ZP2 at 0.95 A resolution | | Descriptor: | Zona pellucida sperm-binding protein 2 | | Authors: | Dioguardi, E, Han, L, Nishimura, K, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
5IIB
 
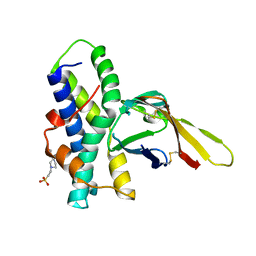 | | Crystal structure of red abalone egg VERL repeat 3 in complex with sperm lysin at 1.64 A resolution (crystal form II) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Egg-lysin, ... | | Authors: | Raj, I, Sadat Al-Hosseini, H, Nishimura, K, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
5II5
 
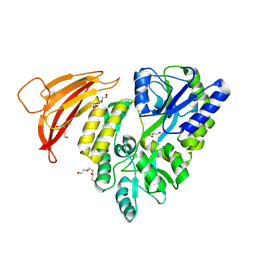 | | Crystal structure of red abalone VERL repeat 1 at 1.8 A resolution | | Descriptor: | Maltose-binding periplasmic protein,Vitelline envelope sperm lysin receptor, TRIETHYLENE GLYCOL, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Sadat Al-Hosseini, H, Raj, I, Nishimura, K, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
5II7
 
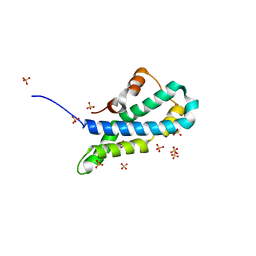 | | In-house sulfur-SAD structure of orthorhombic red abalone lysin at 1.66 A resolution | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Egg-lysin, SULFATE ION | | Authors: | Sadat Al-Hosseini, H, Raj, I, Nishimura, K, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
5II9
 
 | |
5II8
 
 | | Orthorhombic crystal structure of red abalone lysin at 0.99 A resolution | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Egg-lysin, SULFATE ION | | Authors: | Sadat Al-Hosseini, H, Raj, I, Nishimura, K, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
5IIA
 
 | | Crystal structure of red abalone egg VERL repeat 3 in complex with sperm lysin at 1.7 A resolution (crystal form I) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Egg-lysin, Vitelline envelope sperm lysin receptor | | Authors: | Sadat Al-Hosseini, H, Raj, I, Nishimura, K, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
5IIC
 
 | | Crystal structure of red abalone VERL repeat 3 at 2.9 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Maltose-binding periplasmic protein,Vitelline envelope sperm lysin receptor, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Sadat Al-Hosseini, H, Raj, I, Nishimura, K, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
1MYQ
 
 | | An intramolecular quadruplex of (GGA)(4) triplet repeat DNA with a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad, and its dimeric interaction | | Descriptor: | 5'-D(*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*GP*A)-3' | | Authors: | Matsugami, A, Ouhashi, K, Kanagawa, M, Liu, H, Kanagawa, S, Uesugi, S, Katahira, M. | | Deposit date: | 2002-10-04 | | Release date: | 2002-10-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An intramolecular quadruplex of (GGA)(4) triplet repeat DNA with a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad, and its dimeric interaction.
J.Mol.Biol., 313, 2001
|
|
1X0R
 
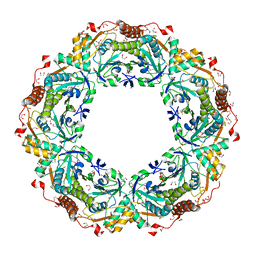 | | Thioredoxin Peroxidase from Aeropyrum pernix K1 | | Descriptor: | 1,2-ETHANEDIOL, Probable peroxiredoxin | | Authors: | Nakamura, T, Yamamoto, T, Inoue, T, Matsumura, H, Kobayashi, A, Hagihara, Y, Uegaki, K, Ataka, M, Kai, Y, Ishikawa, K. | | Deposit date: | 2005-03-28 | | Release date: | 2005-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of thioredoxin peroxidase from aerobic hyperthermophilic archaeon Aeropyrum pernix K1
Proteins, 62, 2006
|
|
7VUS
 
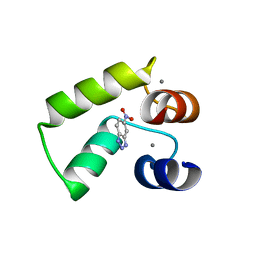 | | Crystal structure of AlleyCat9 with 5-nitro-benzotriazole | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-nitro-1H-benzotriazole, AlleyCat, ... | | Authors: | Tame, J.R.H, Korendovych, I.V, Margheritis, E, Takahashi, K. | | Deposit date: | 2021-11-04 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | NMR-guided directed evolution.
Nature, 610, 2022
|
|
7VUR
 
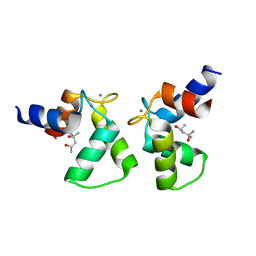 | | Crystal structure of AlleyCat9 with calcium but no inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, AlleyCat, CALCIUM ION | | Authors: | Margheritis, E, Takahashi, K, Korendovych, I.V, Tame, J.R.H. | | Deposit date: | 2021-11-04 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | NMR-guided directed evolution.
Nature, 610, 2022
|
|
7VUU
 
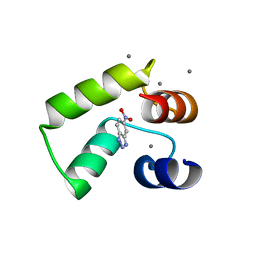 | | Crystal structure of AlleyCat10 with inhibitor | | Descriptor: | 5-nitro-1H-benzotriazole, AlleyCat, CALCIUM ION | | Authors: | Tame, J.R.H, Korendovych, I.V, Margheritis, E, Takahashi, K. | | Deposit date: | 2021-11-04 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | NMR-guided directed evolution.
Nature, 610, 2022
|
|
7VUT
 
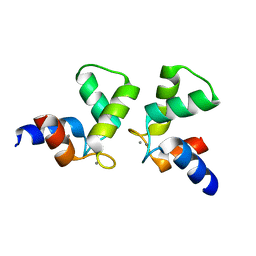 | | Crystal structure of AlleyCat10 | | Descriptor: | AlleyCat10, CALCIUM ION | | Authors: | Tame, J.R.H, Korendovych, I.V, Margheritis, E, Takahashi, K. | | Deposit date: | 2021-11-04 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | NMR-guided directed evolution.
Nature, 610, 2022
|
|
7WWX
 
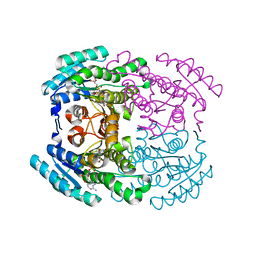 | | Crystal structure of Herbaspirillum huttiense L-arabinose 1-dehydrogenase (NAD bound form) | | Descriptor: | DI(HYDROXYETHYL)ETHER, NAD(P)-dependent dehydrogenase (Short-subunit alcohol dehydrogenase family), NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Matsubara, R, Yoshiwara, K, Watanabe, Y, Watanabe, S. | | Deposit date: | 2022-02-14 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structure of L-arabinose 1-dehydrogenase as a short-chain reductase/dehydrogenase protein.
Biochem.Biophys.Res.Commun., 604, 2022
|
|
3VMZ
 
 | |
7KPW
 
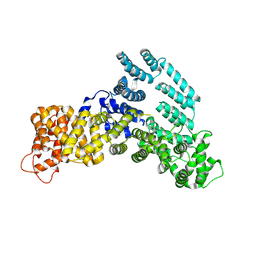 | | Structure of the H-lobe of yeast CKM | | Descriptor: | Mediator of RNA polymerase II transcription subunit 12 | | Authors: | Li, Y.C, Chao, T.C, Kim, H.J, Cholko, T, Chen, S.F, Nakanishi, K, Chang, C.E, Murakami, K, Garcia, B.A, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structure and noncanonical Cdk8 activation mechanism within an Argonaute-containing Mediator kinase module.
Sci Adv, 7, 2021
|
|
3T37
 
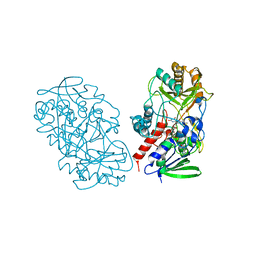 | | Crystal structure of pyridoxine 4-oxidase from Mesorbium loti | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Probable dehydrogenase | | Authors: | Mugo, A.N, Kobayashi, J, Mikami, B, Ohnishi, K, Yagi, T. | | Deposit date: | 2011-07-25 | | Release date: | 2012-08-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Structure biology and crystallization communication
TO BE PUBLISHED
|
|
3VMY
 
 | |
7KPV
 
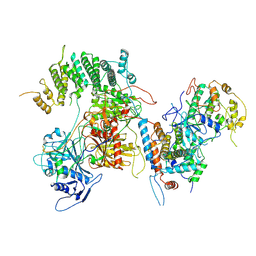 | | Structure of kinase and Central lobes of yeast CKM | | Descriptor: | Mediator of RNA polymerase II transcription subunit 12, Mediator of RNA polymerase II transcription subunit 13, Meiotic mRNA stability protein kinase SSN3, ... | | Authors: | Li, Y.C, Chao, T.C, Kim, H.J, Cholko, T, Chen, S.F, Nakanishi, K, Chang, C.E, Murakami, K, Garcia, B.A, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and noncanonical Cdk8 activation mechanism within an Argonaute-containing Mediator kinase module.
Sci Adv, 7, 2021
|
|
7EFC
 
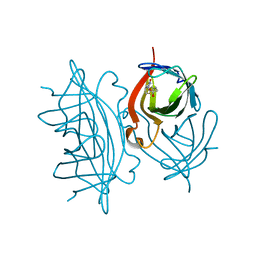 | | 1.70 A cryo-EM structure of streptavidin | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Kotecha, A, Nureki, O. | | Deposit date: | 2021-03-21 | | Release date: | 2021-04-28 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (1.7 Å) | | Cite: | 1.70 A cryo-EM structure of streptavidin using all frames (corresponding to 70 e/A^2 total dose)
To Be Published
|
|
7EFD
 
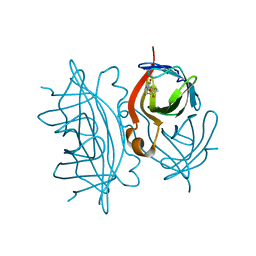 | | 1.77 A cryo-EM structure of Streptavidin using first 40 frames (corresponding to about 40 e/A^2 total dose) | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Kotecha, A, Nureki, O. | | Deposit date: | 2021-03-21 | | Release date: | 2021-04-28 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (1.77 Å) | | Cite: | 1.77 A cryo-EM structure of Streptavidin using first 40 frames (corresponding to about 40e/A^2 total dose)
To Be Published
|
|
