7KUI
 
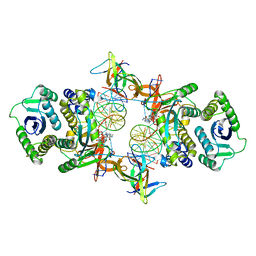 | | Cryo-EM structure of Rous sarcoma virus cleaved synaptic complex (CSC) with HIV-1 integrase strand transfer inhibitor MK-2048. CIC region of a cluster identified by 3-dimensional variability analysis in cryoSPARC. | | Descriptor: | (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-1,9-dioxo-1,2,6,7,8,9-hexahydropyrazino[1',2':1,5]pyrrolo[2,3-d]pyridazine-4-carboxamide, DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*T)-3'), DNA (5'-D(*AP*TP*TP*GP*CP*AP*TP*AP*AP*GP*AP*CP*AP*AP*CP*A)-3'), ... | | Authors: | Pandey, K.K, Bera, S, Shi, K, Aihara, H, Grandgenett, D.P. | | Deposit date: | 2020-11-25 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the Rous sarcoma virus octameric cleaved synaptic complex intasome.
Commun Biol, 4, 2021
|
|
7KU7
 
 | | Cryo-EM structure of Rous sarcoma virus cleaved synaptic complex (CSC) with HIV-1 integrase strand transfer inhibitor MK-2048. Cluster identified by 3-dimensional variability analysis in cryoSPARC. | | Descriptor: | (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-1,9-dioxo-1,2,6,7,8,9-hexahydropyrazino[1',2':1,5]pyrrolo[2,3-d]pyridazine-4-carboxamide, DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*T)-3'), DNA (5'-D(*AP*TP*TP*GP*CP*AP*TP*AP*AP*GP*AP*CP*AP*AP*CP*A)-3'), ... | | Authors: | Pandey, K.K, Bera, S, Shi, K, Aihara, H, Grandgenett, D.P. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the Rous sarcoma virus octameric cleaved synaptic complex intasome.
Commun Biol, 4, 2021
|
|
7KII
 
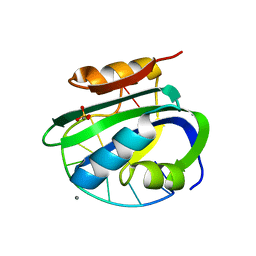 | | Muscovy duck circovirus Rep domain complexed with a single-stranded DNA 10-mer comprising the cleavage site and Mn2+ | | Descriptor: | ATP-dependent helicase Rep, CALCIUM ION, DNA (5'-D(*TP*AP*TP*TP*AP*TP*TP*AP*CP*C)-3'), ... | | Authors: | Tompkins, K.J, Gordon, W.R, Shi, K. | | Deposit date: | 2020-10-23 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Watson-Crick Base-Pairing Requirements for ssDNA Recognition and Processing in Replication-Initiating HUH Endonucleases.
Mbio, 14, 2023
|
|
7KIJ
 
 | | Muscovy duck circovirus Rep domain complexed with a single-stranded DNA 10-mer comprising the cleavage site | | Descriptor: | ATP-dependent helicase Rep, DNA (5'-D(*TP*AP*TP*TP*AP*TP*TP*AP*CP*C)-3'), GLYCEROL, ... | | Authors: | Tompkins, K.J, Gordon, W.R, Shi, K. | | Deposit date: | 2020-10-23 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Watson-Crick Base-Pairing Requirements for ssDNA Recognition and Processing in Replication-Initiating HUH Endonucleases.
Mbio, 14, 2023
|
|
7KIK
 
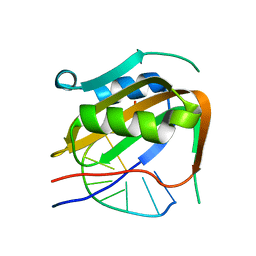 | | Wheat dwarf virus Rep domain circular permutation complexed with a single-stranded DNA 10-mer comprising the cleavage site and Mn2+ | | Descriptor: | DNA (5'-D(*TP*AP*AP*TP*AP*TP*TP*AP*CP*C)-3'), MANGANESE (II) ION, Replication-associated protein | | Authors: | Tompkins, K.J, Gordon, W.R, Shi, K, Litzau, L.A. | | Deposit date: | 2020-10-23 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Watson-Crick Base-Pairing Requirements for ssDNA Recognition and Processing in Replication-Initiating HUH Endonucleases.
Mbio, 14, 2023
|
|
7KM6
 
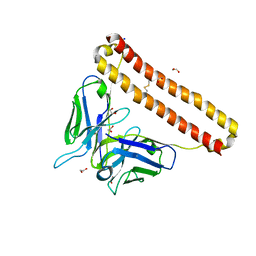 | | APOBEC3B antibody 5G7 Fv-clasp | | Descriptor: | 1,2-ETHANEDIOL, 5G7 human monoclonal FAB heavy chain, 5G7 human monoclonal FAB light chain, ... | | Authors: | Tang, H, Shi, K, Aihara, H. | | Deposit date: | 2020-11-02 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural Characterization of a Minimal Antibody against Human APOBEC3B.
Viruses, 13, 2021
|
|
7KM5
 
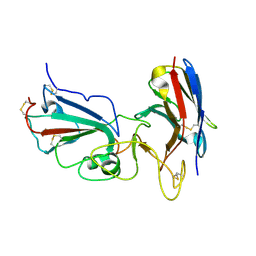 | | Crystal structure of SARS-CoV-2 RBD complexed with Nanosota-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Spike protein S1, ... | | Authors: | Ye, G, Shi, K, Aihara, H, Li, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The development of Nanosota - 1 as anti-SARS-CoV-2 nanobody drug candidates.
Elife, 10, 2021
|
|
6LPK
 
 | | A2AR crystallized in EROCOC17+4, LCP-SFX at 293 K | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Ihara, K, Hato, M, Nakane, T, Yamashita, K, Kimura-Someya, T, Hosaka, T, Ishizuka-Katsura, Y, Tanaka, R, Tanaka, T, Sugahara, M, Hirata, K, Yamamoto, M, Nureki, O, Tono, K, Nango, E, Iwata, S, Shirouzu, M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isoprenoid-chained lipid EROCOC 17+4 : a new matrix for membrane protein crystallization and a crystal delivery medium in serial femtosecond crystallography.
Sci Rep, 10, 2020
|
|
6LPJ
 
 | | A2AR crystallized in EROCOC17+4, LCP-SFX at 277 K | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Ihara, K, Hato, M, Nakane, T, Yamashita, K, Kimura-Someya, T, Hosaka, T, Ishizuka-Katsura, Y, Tanaka, R, Tanaka, T, Sugahara, M, Hirata, K, Yamamoto, M, Nureki, O, Tono, K, Nango, E, Iwata, S, Shirouzu, M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isoprenoid-chained lipid EROCOC 17+4 : a new matrix for membrane protein crystallization and a crystal delivery medium in serial femtosecond crystallography.
Sci Rep, 10, 2020
|
|
6LPL
 
 | | A2AR crystallized in EROCOC17+4, SS-ROX at 100 K | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Ihara, K, Hato, M, Nakane, T, Yamashita, K, Kimura-Someya, T, Hosaka, T, Ishizuka-Katsura, Y, Tanaka, R, Tanaka, T, Sugahara, M, Hirata, K, Yamamoto, M, Nureki, O, Tono, K, Nango, E, Iwata, S, Shirouzu, M. | | Deposit date: | 2020-01-11 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Isoprenoid-chained lipid EROCOC 17+4 : a new matrix for membrane protein crystallization and a crystal delivery medium in serial femtosecond crystallography.
Sci Rep, 10, 2020
|
|
1QCM
 
 | | AMYLOID BETA PEPTIDE (25-35), NMR, 20 STRUCTURES | | Descriptor: | AMYLOID BETA PEPTIDE | | Authors: | Kohno, T, Kobayashi, K, Maeda, T, Sato, K, Takashima, A. | | Deposit date: | 1996-07-19 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structures of the amyloid beta peptide (25-35) in membrane-mimicking environment.
Biochemistry, 35, 1996
|
|
5WQC
 
 | | Crystal structure of human orexin 2 receptor bound to the selective antagonist EMPA determined by the synchrotron light source at SPring-8. | | Descriptor: | N-ethyl-2-[(6-methoxypyridin-3-yl)-(2-methylphenyl)sulfonyl-amino]-N-(pyridin-3-ylmethyl)ethanamide, OLEIC ACID, Orexin receptor type 2,GlgA glycogen synthase,Orexin receptor type 2, ... | | Authors: | Suno, R, Hirata, K, Yamashita, K, Tsujimoto, H, Sasanuma, M, Horita, S, Yamamoto, M, Rosenbaum, D.M, Iwata, S, Shimamura, T, Kobayashi, T. | | Deposit date: | 2016-11-25 | | Release date: | 2017-11-29 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structures of Human Orexin 2 Receptor Bound to the Subtype-Selective Antagonist EMPA
Structure, 26, 2018
|
|
8J9F
 
 | | Structure of STG-hydrolyzing beta-glucosidase 1 (PSTG1) | | Descriptor: | Beta-glucosidase, GLYCEROL | | Authors: | Yanai, T, Imaizumi, R, Takahashi, Y, Katsumura, E, Yamamoto, M, Nakayama, T, Yamashita, S, Takeshita, K, Sakai, N, Matsuura, H. | | Deposit date: | 2023-05-03 | | Release date: | 2024-04-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into a bacterial beta-glucosidase capable of degrading sesaminol triglucoside to produce sesaminol: toward the understanding of the aglycone recognition mechanism by the C-terminal lid domain.
J.Biochem., 174, 2023
|
|
6LMU
 
 | | Cryo-EM structure of the human CALHM2 | | Descriptor: | Calcium homeostasis modulator protein 2 | | Authors: | Demura, K, Kusakizako, T, Shihoya, W, Hiraizumi, M, Shimada, H, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of calcium homeostasis modulator channels in diverse oligomeric assemblies.
Sci Adv, 6, 2020
|
|
6B0B
 
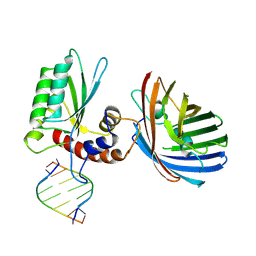 | | Crystal structure of human APOBEC3H | | Descriptor: | APOBEC3H, MCherry, RNA (5'-R(*UP*AP*AP*AP*AP*AP*AP*A)-3'), ... | | Authors: | Shaban, N.M, Shi, K, Banerjee, S, Harris, R.S, Aihara, H. | | Deposit date: | 2017-09-14 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2800622 Å) | | Cite: | The Antiviral and Cancer Genomic DNA Deaminase APOBEC3H Is Regulated by an RNA-Mediated Dimerization Mechanism.
Mol. Cell, 69, 2018
|
|
6LMX
 
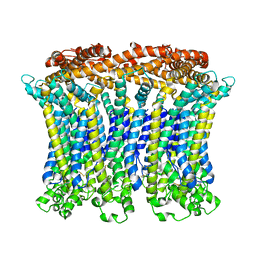 | | Cryo-EM structure of the CALHM chimeric construct (9-mer) | | Descriptor: | Calcium homeostasis modulator 1,Calcium homeostasis modulator protein 2 | | Authors: | Demura, K, Kusakizako, T, Shihoya, W, Hiraizumi, M, Shimada, H, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of calcium homeostasis modulator channels in diverse oligomeric assemblies.
Sci Adv, 6, 2020
|
|
6LMV
 
 | | Cryo-EM structure of the C. elegans CLHM-1 | | Descriptor: | Calcium homeostasis modulator protein | | Authors: | Demura, K, Kusakizako, T, Shihoya, W, Hiraizumi, M, Shimada, H, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of calcium homeostasis modulator channels in diverse oligomeric assemblies.
Sci Adv, 6, 2020
|
|
6LMW
 
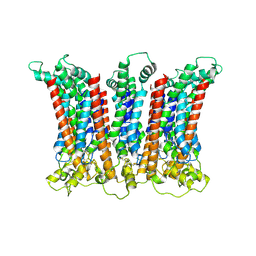 | | Cryo-EM structure of the CALHM chimeric construct (8-mer) | | Descriptor: | Calcium homeostasis modulator 1,Calcium homeostasis modulator protein 2 | | Authors: | Demura, K, Kusakizako, T, Shihoya, W, Hiraizumi, M, Shimada, H, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of calcium homeostasis modulator channels in diverse oligomeric assemblies.
Sci Adv, 6, 2020
|
|
5JGB
 
 | | Crystal structure of human TAK1/TAB1 fusion protein in complex with ligand 10 | | Descriptor: | N-(2-methoxy-4-{[3-(4-methylpiperazin-1-yl)propyl]carbamoyl}phenyl)-4-oxo-3,4-dihydrothieno[3,2-d]pyrimidine-7-carboxamide, TAK1 kinase - TAB1 chimera fusion protein | | Authors: | Irie, M, Nakamura, M, Fukami, T.A, Matsuura, T, Morishima, K. | | Deposit date: | 2016-04-20 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of a potent and highly selective transforming growth factor beta receptor-associated kinase 1 (TAK1) inhibitor by structure based drug design (SBDD)
Bioorg.Med.Chem., 24, 2016
|
|
5JGD
 
 | | Crystal structure of human TAK1/TAB1 fusion protein in complex with ligand 12 | | Descriptor: | N-(2-isopropoxy-3-(4-methylpiperazine-1-carbonyl)phenyl)-4-oxo-3,4-dihydrothieno[3,2-d]pyrimidine-7-carboxamide, TAK1 kinase - TAB1 chimera fusion protein | | Authors: | Irie, M, Nakamura, M, Fukami, T.A, Matsuura, T, Morishima, K. | | Deposit date: | 2016-04-20 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Discovery of a potent and highly selective transforming growth factor beta receptor-associated kinase 1 (TAK1) inhibitor by structure based drug design (SBDD)
Bioorg.Med.Chem., 24, 2016
|
|
7Y51
 
 | | Acetylxylan esterase from Caldanaerobacter subterraneus subsp. tengcongensis TTE0866 delta100 mutant | | Descriptor: | GLYCEROL, NICKEL (II) ION, Predicted xylanase/chitin deacetylase | | Authors: | Sasamoto, K, Himiyama, T, Moriyoshi, K, Ohmoto, T, Uegaki, K, Nakamura, T, Nishiya, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Functional analysis of the N-terminal region of acetylxylan esterase from Caldanaerobacter subterraneus subsp. tengcongensis.
Febs Open Bio, 12, 2022
|
|
5KNC
 
 | | Crystal structure of the 3 ADP-bound V1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Mizutani, K, Maruyama, S, Shimono, K, Imai, F.L, Muneyuki, E, Kakinuma, Y, Ishizuka-Katsura, Y, Shirouzu, M, Yokoyama, S, Yamato, I, Murata, T. | | Deposit date: | 2016-06-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.015 Å) | | Cite: | Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor
Nat Commun, 7, 2016
|
|
5KNB
 
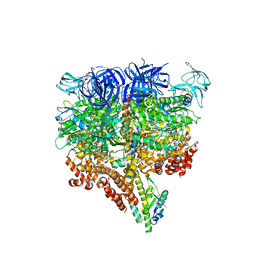 | | Crystal structure of the 2 ADP-bound V1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Mizutani, K, Maruyama, S, Shimono, K, Imai, F.L, Muneyuki, E, Kakinuma, Y, Ishizuka-Katsura, Y, Shirouzu, M, Yokoyama, S, Yamato, I, Murata, T. | | Deposit date: | 2016-06-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.251 Å) | | Cite: | Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor
Nat Commun, 7, 2016
|
|
5KND
 
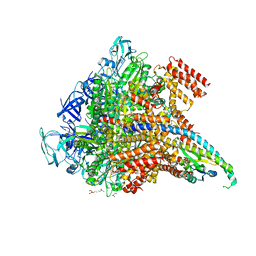 | | Crystal structure of the Pi-bound V1 complex | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Mizutani, K, Maruyama, S, Shimono, K, Imai, F.L, Muneyuki, E, Kakinuma, Y, Ishizuka-Katsura, Y, Shirouzu, M, Yokoyama, S, Yamato, I, Murata, T. | | Deposit date: | 2016-06-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.888 Å) | | Cite: | Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor
Nat Commun, 7, 2016
|
|
3A8R
 
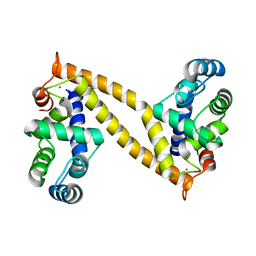 | | The structure of the N-terminal regulatory domain of a plant NADPH oxidase | | Descriptor: | CALCIUM ION, Putative uncharacterized protein | | Authors: | Oda, T, Hashimoto, H, Kuwabara, N, Akashi, S, Hayashi, K, Kojima, C, Wong, H.L, Kawasaki, T, Shimamoto, K, Sato, M, Shimizu, T. | | Deposit date: | 2009-10-07 | | Release date: | 2009-10-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of the N-terminal regulatory domain of a plant NADPH oxidase and its functional implications
J.Biol.Chem., 285, 2010
|
|
