1WMR
 
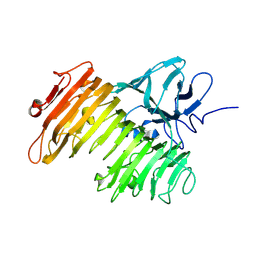 | | Crystal Structure of Isopullulanase from Aspergillus niger ATCC 9642 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isopullulanase | | Authors: | Mizuno, M, Tonozuka, T, Miyasaka, Y, Akeboshi, H, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2004-07-15 | | Release date: | 2005-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Aspergillus niger Isopullulanase, a Member of Glycoside Hydrolase Family 49
J.Mol.Biol., 376, 2008
|
|
1X0C
 
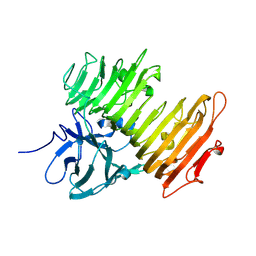 | | Improved Crystal Structure of Isopullulanase from Aspergillus niger ATCC 9642 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isopullulanase | | Authors: | Mizuno, M, Tonozuka, T, Yamamura, A, Miyasaka, Y, Akeboshi, H, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2005-03-17 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Aspergillus niger Isopullulanase, a Member of Glycoside Hydrolase Family 49
J.Mol.Biol., 376, 2008
|
|
6A0R
 
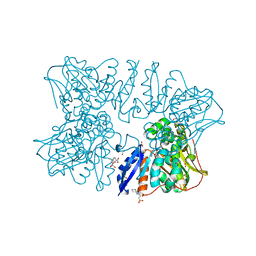 | | Homoserine dehydrogenase from Thermus thermophilus HB8 unliganded form | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Akai, S, Ikushiro, H, Sawai, T, Yano, T, Kamiya, N, Miyahara, I. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The crystal structure of homoserine dehydrogenase complexed with l-homoserine and NADPH in a closed form
J. Biochem., 165, 2019
|
|
6A0S
 
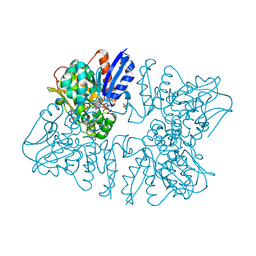 | | Homoserine dehydrogenase from Thermus thermophilus HB8 complexed with HSE and NADPH | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Akai, S, Ikushiro, H, Sawai, T, Yano, T, Kamiya, N, Miyahara, I. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of homoserine dehydrogenase complexed with l-homoserine and NADPH in a closed form
J. Biochem., 165, 2019
|
|
3VJM
 
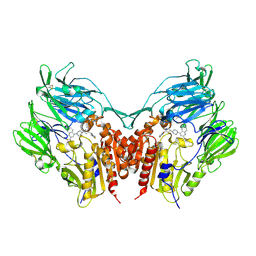 | | Crystal structure of human depiptidyl peptidase IV (DPP-4) in complex with a prolylthiazolidine inhibitor #1 | | Descriptor: | 1,3-thiazolidin-3-yl[(2S,4S)-4-{4-[2-(trifluoromethyl)quinolin-4-yl]piperazin-1-yl}pyrrolidin-2-yl]methanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Akahoshi, F, Kishida, H, Miyaguchi, I, Yoshida, T, Ishii, S. | | Deposit date: | 2011-10-24 | | Release date: | 2012-08-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fused bicyclic heteroarylpiperazine-substituted l-prolylthiazolidines as highly potent DPP-4 inhibitors lacking the electrophilic nitrile group
Bioorg.Med.Chem., 20, 2012
|
|
6A0U
 
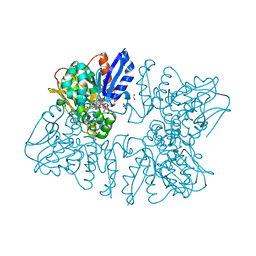 | | Homoserine dehydrogenase K195A mutant from Thermus thermophilus HB8 complexed with HSE and NADP+ | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Akai, S, Ikushiro, H, Sawai, T, Yano, T, Kamiya, N, Miyahara, I. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The crystal structure of homoserine dehydrogenase complexed with l-homoserine and NADPH in a closed form
J. Biochem., 165, 2019
|
|
6A0T
 
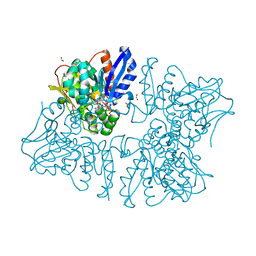 | | Homoserine dehydrogenase K99A mutant from Thermus thermophilus HB8 complexed with HSE and NADP+ | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Akai, S, Ikushiro, H, Sawai, T, Yano, T, Kamiya, N, Miyahara, I. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The crystal structure of homoserine dehydrogenase complexed with l-homoserine and NADPH in a closed form
J. Biochem., 165, 2019
|
|
3UES
 
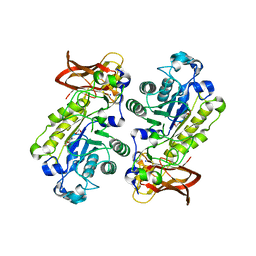 | | Crystal structure of alpha-1,3/4-fucosidase from Bifidobacterium longum subsp. infantis complexed with deoxyfuconojirimycin | | Descriptor: | (2S,3R,4S,5R)-2-METHYLPIPERIDINE-3,4,5-TRIOL, 1,2-ETHANEDIOL, Alpha-1,3/4-fucosidase, ... | | Authors: | Sakurama, H, Fushinobu, S, Yoshida, E, Honda, Y, Hidaka, M, Ashida, H, Kitaoka, M, Katayama, T, Yamamoto, K, Kumagai, H. | | Deposit date: | 2011-10-31 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1,3-1,4-alpha-L-fucosynthase that specifically introduces Lewis a/x antigens into type-1/2 chains
J.Biol.Chem., 287, 2012
|
|
3UET
 
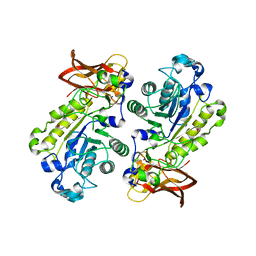 | | Crystal structure of alpha-1,3/4-fucosidase from Bifidobacterium longum subsp. infantis D172A/E217A mutant complexed with lacto-N-fucopentaose II | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,3/4-fucosidase, SODIUM ION, ... | | Authors: | Sakurama, H, Fushinobu, S, Yoshida, E, Honda, Y, Hidaka, M, Ashida, H, Kitaoka, M, Katayama, T, Yamamoto, K, Kumagai, H. | | Deposit date: | 2011-10-31 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 1,3-1,4-alpha-L-fucosynthase that specifically introduces Lewis a/x antigens into type-1/2 chains
J.Biol.Chem., 287, 2012
|
|
3W39
 
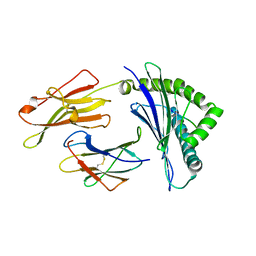 | | Crystal structure of HLA-B*5201 in complexed with HIV immunodominant epitope (TAFTIPSI) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-52 alpha chain, ... | | Authors: | Yagita, Y, Kuse, N, Kuroki, K, Gatanaga, H, Carlson, J.M, Chikata, T, Brumme, Z.L, Murakoshi, H, Akahoshi, T, Pfeifer, N, Mallal, S, John, M, Ose, T, Matsubara, H, Kanda, R, Fukunaga, Y, Honda, K, Kawashima, Y, Ariumi, Y, Oka, S, Maenaka, K, Takiguchi, M. | | Deposit date: | 2012-12-13 | | Release date: | 2013-02-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Distinct HIV-1 Escape Patterns Selected by Cytotoxic T Cells with Identical Epitope Specificity
J.Virol., 87, 2013
|
|
3WYY
 
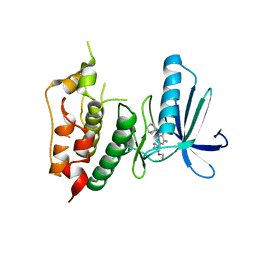 | | CRYSTAL STRUCTURE OF HUMAN MPS1 CATALYTIC DOMAIN IN COMPLEX WITH (E)-3-(4-((6-(((3s,5s,7s)-adamantan-1-yl)amino)-4-amino-5-cyanopyridin-2-yl)amino)-2-(cyanomethoxy)phenyl)-N-(2-methoxyethyl)acrylamide | | Descriptor: | (2E)-3-[4-({4-amino-5-cyano-6-[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]dec-1-ylamino]pyridin-2-yl}amino)-2-(cyanomethoxy)phenyl]-N-(2-methoxyethyl)prop-2-enamide, Dual specificity protein kinase TTK | | Authors: | Kusakabe, K, Ide, N, Daigo, Y, Itoh, T, Yamamoto, T, Kojima, E, Mitsuoka, Y, Tadano, G, Tagashira, S, Higashino, K, Okano, Y, Sato, Y, Inoue, M, Iguchi, M, Kanazawa, T, Ishioka, Y, Dohi, K, Kido, Y, Sakamoto, S, Ando, S, Maeda, M, Higaki, M, Yoshizawa, H, Murai, H, Nakamura, Y. | | Deposit date: | 2014-09-10 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | A unique hinge binder of extremely selective aminopyridine-based Mps1 (TTK) kinase inhibitors with cellular activity.
Bioorg.Med.Chem., 23, 2015
|
|
1WDA
 
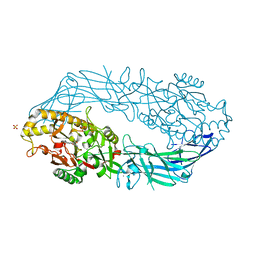 | | Crystal structure of human peptidylarginine deiminase type4 (PAD4) in complex with benzoyl-L-arginine amide | | Descriptor: | CALCIUM ION, N-[(E)-2-AMINO-1-(3-{[AMINO(IMINO)METHYL]AMINO}PROPYL)-2-HYDROXYVINYL]BENZAMIDE, Protein-arginine deiminase type IV, ... | | Authors: | Arita, K, Hashimoto, H, Shimizu, T, Nakashima, K, Yamada, M, Sato, M. | | Deposit date: | 2004-05-12 | | Release date: | 2004-07-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for Ca(2+)-induced activation of human PAD4
Nat.Struct.Mol.Biol., 11, 2004
|
|
1WD9
 
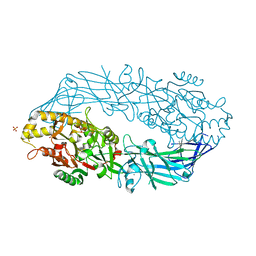 | | Calcium bound form of human peptidylarginine deiminase type4 (PAD4) | | Descriptor: | CALCIUM ION, Protein-arginine deiminase type IV, SULFATE ION | | Authors: | Arita, K, Hashimoto, H, Shimizu, T, Nakashima, K, Yamada, M, Sato, M. | | Deposit date: | 2004-05-12 | | Release date: | 2004-07-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for Ca(2+)-induced activation of human PAD4
Nat.Struct.Mol.Biol., 11, 2004
|
|
1WD8
 
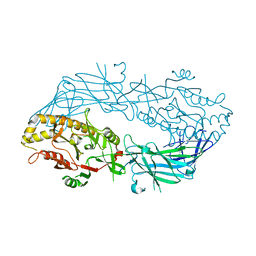 | | Calcium free form of human peptidylarginine deiminase type4 (PAD4) | | Descriptor: | Protein-arginine deiminase type IV | | Authors: | Arita, K, Hashimoto, H, Shimizu, T, Nakashima, K, Yamada, M, Sato, M. | | Deposit date: | 2004-05-12 | | Release date: | 2004-07-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for Ca(2+)-induced activation of human PAD4
Nat.Struct.Mol.Biol., 11, 2004
|
|
3WO4
 
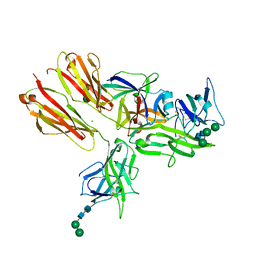 | | Crystal structure of the IL-18 signaling ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tsutsumi, N, Kimura, T, Arita, K, Ariyoshi, M, Ohnishi, H, Kondo, N, Shirakawa, M, Kato, Z, Tochio, H. | | Deposit date: | 2013-12-19 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis for receptor recognition of human interleukin-18
Nat Commun, 5, 2014
|
|
3WO2
 
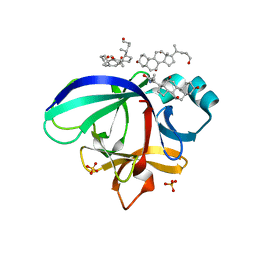 | | Crystal structure of human interleukin-18 | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Interleukin-18, SULFATE ION | | Authors: | Tsutsumi, N, Kimura, T, Arita, K, Ariyoshi, M, Ohnishi, H, Kondo, N, Shirakawa, M, Kato, Z, Tochio, H. | | Deposit date: | 2013-12-19 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The structural basis for receptor recognition of human interleukin-18
Nat Commun, 5, 2014
|
|
3WO3
 
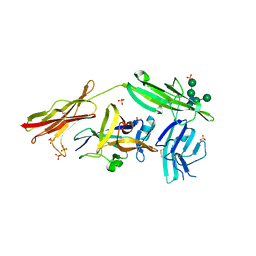 | | Crystal structure of IL-18 in complex with IL-18 receptor alpha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tsutsumi, N, Kimura, T, Arita, K, Ariyoshi, M, Ohnishi, H, Kondo, N, Shirakawa, M, Kato, Z, Tochio, H. | | Deposit date: | 2013-12-19 | | Release date: | 2014-12-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis for receptor recognition of human interleukin-18
Nat Commun, 5, 2014
|
|
2D0S
 
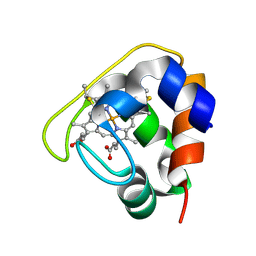 | | Crystal structure of the Cytochrome C552 from moderate thermophilic bacterium, hydrogenophilus thermoluteolus | | Descriptor: | HEME C, cytochrome c | | Authors: | Nakamura, S, Ichiki, S.I, Takashima, H, Uchiyama, S, Hasegawa, J, Kobayashi, Y, Sambongi, Y, Ohkubo, T. | | Deposit date: | 2005-08-08 | | Release date: | 2006-05-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Cytochrome c552 from a Moderate Thermophilic Bacterium, Hydrogenophilus thermoluteolus: Comparative Study on the Thermostability of Cytochrome c
Biochemistry, 45, 2006
|
|
5XUV
 
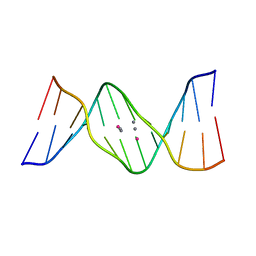 | | Crystal structure of DNA duplex containing 4-thiothymine-2Ag(I)-4-thiothymine base pairs | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*(8RO)P*(8RO)P*TP*CP*GP*CP*G)-3'), POTASSIUM ION, SILVER ION | | Authors: | Kondo, J, Sugawara, T, Saneyoshi, H, Ono, A. | | Deposit date: | 2017-06-26 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a DNA duplex containing four Ag(i) ions in consecutive dinuclear Ag(i)-mediated base pairs: 4-thiothymine-2Ag(i)-4-thiothymine
Chem. Commun. (Camb.), 53, 2017
|
|
5Y34
 
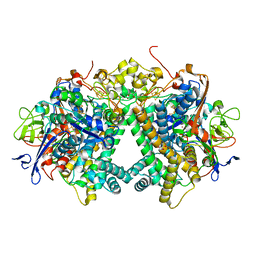 | | Membrane-bound respiratory [NiFe]-hydrogenase from Hydrogenovibrio marinus in a ferricyanide-oxidized condition | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, FE4-S3 CLUSTER, ... | | Authors: | Shomura, Y, Yoon, K.S, Nishihara, H, Higuchi, Y. | | Deposit date: | 2017-07-27 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural basis for a [4Fe-3S] cluster in the oxygen-tolerant membrane-bound [NiFe]-hydrogenase
Nature, 479, 2011
|
|
5ZOX
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH 7 at 288 K (1) | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase, SODIUM ION | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.691 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZP9
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH 6 at 283 K (1) | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase, SODIUM ION | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZPO
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by phenylethylamine at pH 8 at 288 K (2) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, PHENYLACETALDEHYDE, ... | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZP6
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH 6 at 277 K (2) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Phenylethylamine oxidase, ... | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.688 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZPK
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by phenylethylamine at pH 6 at 288 K (2) | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase, SODIUM ION | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
