9C4C
 
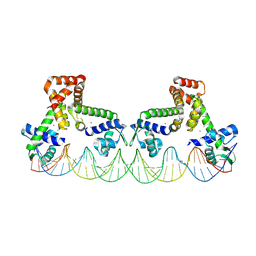 | | The structure of two MntR dimers bound to the native mnep promoter sequence | | Descriptor: | DNA (38-MER), DNA (39-MER), HTH-type transcriptional regulator MntR, ... | | Authors: | Shi, H, Fu, Y, Glasfeld, A, Ahuja, S. | | Deposit date: | 2024-06-04 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | The structure of two MntR dimers bound to the native mnep promoter sequence.
To Be Published
|
|
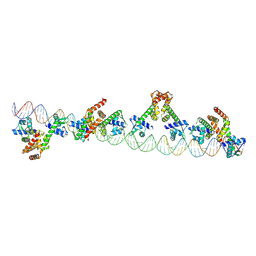
9C4D
 
 | |
8BQN
 
 | |
8F49
 
 | |
8F4L
 
 | |
8EW2
 
 | |
8EW0
 
 | |
8F7Y
 
 | |
5UBV
 
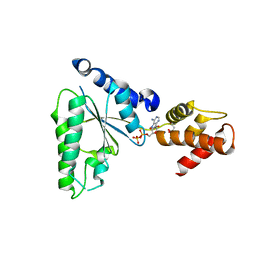 | |
4LL7
 
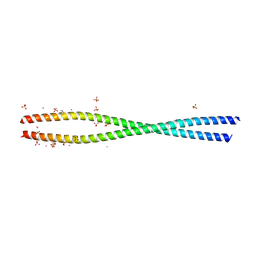 | | Structure of She3p amino terminus. | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DYSPROSIUM ION, ... | | Authors: | Shi, H, Singh, N, Esselborn, F, Blobel, G. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of a myosinbulletadaptor complex and pairing by cargo.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7JU1
 
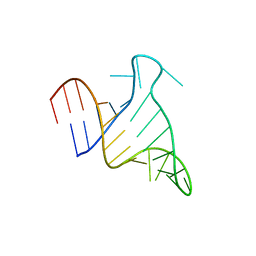 | | The FARFAR-NMR Ensemble of 29-mer HIV-1 Trans-activation Response Element RNA (N=20) | | Descriptor: | RNA (29-MER) | | Authors: | Shi, H, Rangadurai, A, Roy, R, Yesselman, J.D, Al-Hashimi, H.M. | | Deposit date: | 2020-08-18 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rapid and accurate determination of atomistic RNA dynamic ensemble models using NMR and structure prediction
Nat Commun, 11, 2020
|
|
8HHS
 
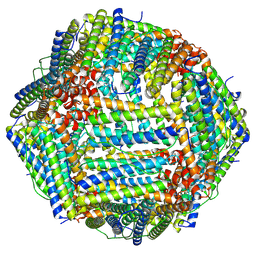 | |
4LL6
 
 | | Structure of Myo4p globular tail domain. | | Descriptor: | ACETIC ACID, Myosin-4 | | Authors: | Shi, H, Singh, N, Esselborn, F, Blobel, G. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a myosinbulletadaptor complex and pairing by cargo.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4LL8
 
 | | Complex of carboxy terminal domain of Myo4p and She3p middle fragment | | Descriptor: | Myosin-4, SWI5-dependent HO expression protein 3 | | Authors: | Shi, H, Singh, N, Esselborn, F, Blobel, G. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.578 Å) | | Cite: | Structure of a myosinbulletadaptor complex and pairing by cargo.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3OPB
 
 | | Crystal structure of She4p | | Descriptor: | SWI5-dependent HO expression protein 4 | | Authors: | Shi, H, Blobel, G. | | Deposit date: | 2010-08-31 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | UNC-45/CRO1/She4p (UCS) protein forms elongated dimer and joins two myosin heads near their actin binding region.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8HI2
 
 | |
1OO0
 
 | |
1EHZ
 
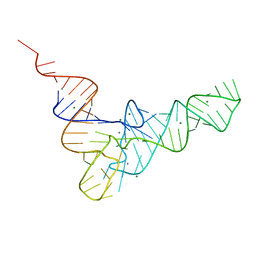 | |
292D
 
 | | INTERACTION BETWEEN THE LEFT-HANDED Z-DNA AND POLYAMINE:THE CRYSTAL STRUCTURE OF THE D(CG)3 AND N-(2-AMINOETHYL)-1,4-DIAMINOBUTANE COMPLEX | | Descriptor: | 1-(AMINOETHYL)AMINO-4-AMINOBUTANE, DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Ohishi, H, Kunisawa, S, Van Der Marel, G, Van Boom, J.H, Rich, A, Wang, A.H.-J, Tomita, K, Hakoshima, T. | | Deposit date: | 1991-10-09 | | Release date: | 1996-12-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Interaction between the left-handed Z-DNA and polyamine. The crystal structure of the d(CG)3 and N-(2-aminoethyl)-1,4-diamino-butane complex.
FEBS Lett., 284, 1991
|
|
1HNR
 
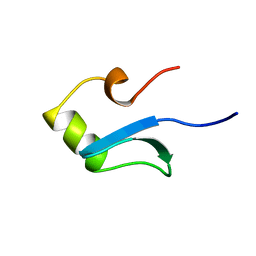 | | H-NS (DNA-BINDING DOMAIN) | | Descriptor: | H-NS | | Authors: | Shindo, H, Iwaki, T, Ieda, R, Kurumizaka, H, Ueguchi, C, Mizuno, T, Morikawa, S, Nakamura, H, Kuboniwa, H. | | Deposit date: | 1995-04-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from Escherichia coli.
FEBS Lett., 360, 1995
|
|
1HNS
 
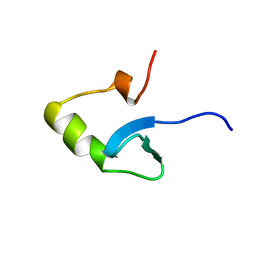 | | H-NS (DNA-BINDING DOMAIN) | | Descriptor: | H-NS | | Authors: | Shindo, H, Iwaki, T, Ieda, R, Kurumizaka, H, Ueguchi, C, Mizuno, T, Morikawa, S, Nakamura, H, Kuboniwa, H. | | Deposit date: | 1995-04-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from Escherichia coli.
FEBS Lett., 360, 1995
|
|
3W08
 
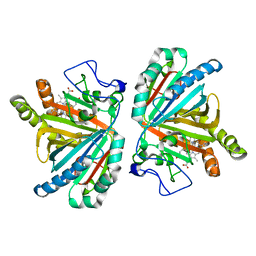 | | Crystal structure of aldoxime dehydratase | | Descriptor: | Aldoxime dehydratase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hashimoto, H, Nomura, J, Hashimoto, Y, Oinuma, K.I, Wada, K, Hishiki, A, Hara, K, Kobayashi, M. | | Deposit date: | 2012-10-24 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of aldoxime dehydratase and its catalytic mechanism involved in carbon-nitrogen triple-bond synthesis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6LGP
 
 | | cryo-EM structure of TRPV3 in lipid nanodisc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, DIUNDECYL PHOSPHATIDYL CHOLINE, Transient receptor potential cation channel subfamily V member 3 | | Authors: | Shimada, H, Kusakizako, T, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-12-05 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structure of lipid nanodisc-reconstituted TRPV3 reveals the gating mechanism.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4XSM
 
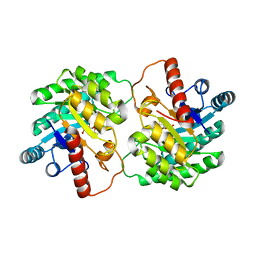 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with D-talitol | | Descriptor: | D-altritol, D-tagatose 3-epimerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
4XSL
 
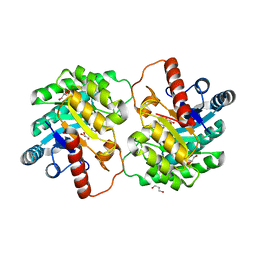 | | Crystal strcutre of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with glycerol | | Descriptor: | D-tagatose 3-epimerase, GLYCEROL, MANGANESE (II) ION | | Authors: | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
