8BQN
 
 | |
7JU1
 
 | | The FARFAR-NMR Ensemble of 29-mer HIV-1 Trans-activation Response Element RNA (N=20) | | Descriptor: | RNA (29-MER) | | Authors: | Shi, H, Rangadurai, A, Roy, R, Yesselman, J.D, Al-Hashimi, H.M. | | Deposit date: | 2020-08-18 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rapid and accurate determination of atomistic RNA dynamic ensemble models using NMR and structure prediction
Nat Commun, 11, 2020
|
|
8F49
 
 | |
8F4L
 
 | |
8EW2
 
 | |
8F7Y
 
 | |
8EW0
 
 | |
3OPB
 
 | | Crystal structure of She4p | | Descriptor: | SWI5-dependent HO expression protein 4 | | Authors: | Shi, H, Blobel, G. | | Deposit date: | 2010-08-31 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | UNC-45/CRO1/She4p (UCS) protein forms elongated dimer and joins two myosin heads near their actin binding region.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8HHS
 
 | |
5UBV
 
 | |
4LL7
 
 | | Structure of She3p amino terminus. | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DYSPROSIUM ION, ... | | Authors: | Shi, H, Singh, N, Esselborn, F, Blobel, G. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of a myosinbulletadaptor complex and pairing by cargo.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8HI2
 
 | |
1OO0
 
 | |
4LL6
 
 | | Structure of Myo4p globular tail domain. | | Descriptor: | ACETIC ACID, Myosin-4 | | Authors: | Shi, H, Singh, N, Esselborn, F, Blobel, G. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a myosinbulletadaptor complex and pairing by cargo.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4LL8
 
 | | Complex of carboxy terminal domain of Myo4p and She3p middle fragment | | Descriptor: | Myosin-4, SWI5-dependent HO expression protein 3 | | Authors: | Shi, H, Singh, N, Esselborn, F, Blobel, G. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-12 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (3.578 Å) | | Cite: | Structure of a myosinbulletadaptor complex and pairing by cargo.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1EHZ
 
 | |
4ZOH
 
 | | Crystal structure of glyceraldehyde oxidoreductase | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Nishimasu, H, Fushinobu, S, Wakagi, T. | | Deposit date: | 2015-05-06 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Archaeal Mo-Containing Glyceraldehyde Oxidoreductase Isozymes Exhibit Diverse Substrate Specificities through Unique Subunit Assemblies.
Plos One, 11, 2016
|
|
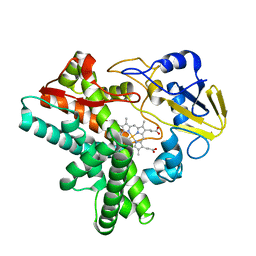
1EHF
 
 | |
4GEM
 
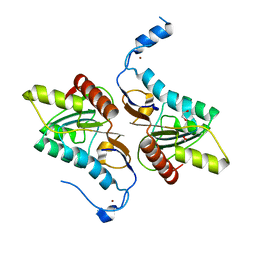 | | Crystal structure of Zucchini (K171A) | | Descriptor: | 1,2-ETHANEDIOL, Mitochondrial cardiolipin hydrolase, ZINC ION | | Authors: | Nishimasu, H, Fukuhara, S, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-02 | | Release date: | 2012-10-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structure and function of Zucchini endoribonuclease in piRNA biogenesis
Nature, 491, 2012
|
|
4GEN
 
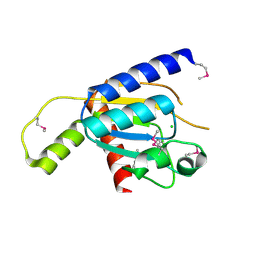 | | Crystal structure of Zucchini (monomer) | | Descriptor: | CHLORIDE ION, Mitochondrial cardiolipin hydrolase | | Authors: | Nishimasu, H, Fukuhara, S, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-02 | | Release date: | 2012-10-17 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of Zucchini endoribonuclease in piRNA biogenesis
Nature, 491, 2012
|
|
4Q0Q
 
 | | Crystal structure of Acinetobacter sp. DL28 L-ribose isomerase in complex with L-ribulose | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), L-Ribose isomerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Teraoka, M, Izumori, K, Kamitori, S. | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate.
Febs J., 281, 2014
|
|
4Q0U
 
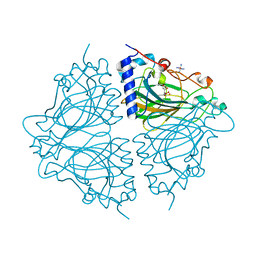 | | Crystal structure of Acinetobacter sp. DL28 L-ribose isomerase mutant E204Q in complex with L-ribose | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), L-Ribose isomerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Teraoka, M, Izumori, K, Kamitori, S. | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate.
Febs J., 281, 2014
|
|
3VKM
 
 | | Protease-resistant mutant form of Human Galectin-8 in complex with sialyllactose and lactose | | Descriptor: | 1,2-ETHANEDIOL, Galectin-8, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Yoshida, H, Yamashita, S, Teraoka, M, Nakakita, S, Nishi, N, Kamitori, S. | | Deposit date: | 2011-11-18 | | Release date: | 2012-09-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | X-ray structure of a protease-resistant mutant form of human galectin-8 with two carbohydrate recognition domains
Febs J., 279, 2012
|
|
5HNW
 
 | | Structural basis of backwards motion in kinesin-14: minus-end directed nKn664 in the AMPPNP state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shigematsu, H, Yokoyama, T, Kikkawa, M, Shirouzu, M, Nitta, R. | | Deposit date: | 2016-01-19 | | Release date: | 2016-08-10 | | Last modified: | 2018-07-25 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural Basis of Backwards Motion in Kinesin-1-Kinesin-14 Chimera: Implication for Kinesin-14 Motility
Structure, 24, 2016
|
|
3VRA
 
 | | Mitochondrial rhodoquinol-fumarate reductase from the parasitic nematode Ascaris suum with the specific inhibitor Atpenin A5 | | Descriptor: | 3-[(2S,4S,5R)-5,6-DICHLORO-2,4-DIMETHYL-1-OXOHEXYL]-4-HYDROXY-5,6-DIMETHOXY-2(1H)-PYRIDINONE, Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Shimizu, H, Shiba, T, Inaoka, D.K, Osanai, A, Kita, K, Sakamoto, K, Harada, S. | | Deposit date: | 2012-04-07 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.44 Å) | | Cite: | Crystal structure of mitochondrial quinol-fumarate reductase from parasitic nematode Ascaris suum
To be Published
|
|
