6KFH
 
 | | Undocked hemichannel of an N-terminal deletion mutant of INX-6 in a nanodisc | | Descriptor: | Innexin-6 | | Authors: | Burendei, B, Shinozaki, R, Watanabe, M, Terada, T, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2019-07-07 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of undocked innexin-6 hemichannels in phospholipids.
Sci Adv, 6, 2020
|
|
7RM5
 
 | | MicroED structure of the human adenosine receptor at 2.8A | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a/Soluble cytochrome b562 chimera, CHOLESTEROL, ... | | Authors: | Martynowycz, M.W, Shiriaeva, A, Ge, X, Hattne, J, Nannenga, B.L, Cherezov, V, Gonen, T. | | Deposit date: | 2021-07-26 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.79 Å) | | Cite: | MicroED structure of the human adenosine receptor determined from a single nanocrystal in LCP.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8SFM
 
 | | Crystal structure of the engineered SsoPox variant IVB10 in alternate state | | Descriptor: | 1,2-ETHANEDIOL, Aryldialkylphosphatase, COBALT (II) ION, ... | | Authors: | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | Deposit date: | 2023-04-11 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
8SFB
 
 | | Crystal structure of the engineered SsoPox variant IVA4 | | Descriptor: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION, ... | | Authors: | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
8SF2
 
 | | Crystal structure of the engineered SsoPox variant IG7 | | Descriptor: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION, ... | | Authors: | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
8SFA
 
 | | Crystal structure of the engineered SsoPox variant IIIC1 | | Descriptor: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION | | Authors: | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
8SF9
 
 | | Crystal structure of the engineered SsoPox variant IG7 - Alternative state | | Descriptor: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION | | Authors: | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
8SFC
 
 | | Crystal structure of the engineered SsoPox variant IVA4 in alternate state | | Descriptor: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION, ... | | Authors: | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
8SFK
 
 | | Crystal structure of the engineered SsoPox variant IVE2 | | Descriptor: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION, ... | | Authors: | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | Deposit date: | 2023-04-11 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
8SFD
 
 | | Crystal structure of the engineered SsoPox variant IVB10 | | Descriptor: | Aryldialkylphosphatase, COBALT (II) ION, FE (III) ION, ... | | Authors: | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Changes in Active Site Loops Conformation Relates to a Transition from Lactonase to Phosphotriesterase
To Be Published
|
|
7REF
 
 | | Structure of MS3494 from Mycobacterium smegmatis | | Descriptor: | BROMIDE ION, MS3494 | | Authors: | Kent, J.E, Aleshin, A.E, Zhang, L, Niederweis, M, Marassi, F.M. | | Deposit date: | 2021-07-12 | | Release date: | 2021-08-18 | | Last modified: | 2022-06-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A periplasmic cinched protein is required for siderophore secretion and virulence of Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
6K52
 
 | | Hyperthermophilic GH6 cellobiohydrolase (HmCel6A) from the microbial flora of a Japanese hot spring | | Descriptor: | ACETATE ION, CALCIUM ION, GH6 cellobiohydrolase, ... | | Authors: | Baba, S, Takeda, M, Okuma, J, Hirose, Y, Nishimura, A, Takata, M, Oda, K, Shibata, D, Kondo, Y, Kumasaka, T. | | Deposit date: | 2019-05-28 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68000138 Å) | | Cite: | A hyperthermophilic cellobiohydrolase mined from a hot spring metagenomic data
To Be Published
|
|
6K53
 
 | | A variant of metagenome-derived GH6 cellobiohydrolase, HmCel6A (P88S/L230F/F414S) | | Descriptor: | CITRATE ANION, GH6 cellobiohydrolase, HMCEL6A, ... | | Authors: | Baba, S, Takeda, M, Okuma, J, Hirose, Y, Nishimura, A, Takata, M, Oda, K, Shibata, D, Kondo, Y, Kumasaka, T. | | Deposit date: | 2019-05-28 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A hyperthermophilic GH6 cellobiohydrolase (HmCel6A) from a hot spring metagenomic data
To Be Published
|
|
6QOY
 
 | | Crystal structure of L1 protease Lysobacter sp. XL1 in complex with AEBSF | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, 4-(2-azanylethyl)benzenesulfonic acid, CHLORIDE ION, ... | | Authors: | Gabdulkhakov, A, Tishchenko, S, Kudryakova, I, Afoshin, A, Vasilyeva, N. | | Deposit date: | 2019-02-13 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Serine bacteriolytic protease L1 of Lysobacter sp. XL1 complexed with protease inhibitor AEBSF: features of interaction
Process Biochem, 2019
|
|
7YA7
 
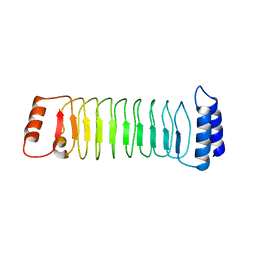 | | The crystal structure of IpaH1.4 LRR domain | | Descriptor: | RING-type E3 ubiquitin transferase | | Authors: | Hiragi, K, Nishide, A, Takagi, K, Iwai, K, Kim, M, Mizushima, T. | | Deposit date: | 2022-06-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insight into the recognition of the linear ubiquitin assembly complex by Shigella E3 ligase IpaH1.4/2.5.
J.Biochem., 173, 2023
|
|
6K54
 
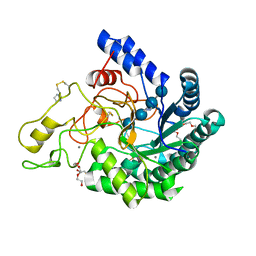 | | Hyperthermophilic GH6 cellobiohydrolase II (HmCel6A) in complex with trisaccharide | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Baba, S, Takeda, M, Okuma, J, Hirose, Y, Nishimura, A, Takata, M, Oda, K, Shibata, D, Kondo, Y, Kumasaka, T. | | Deposit date: | 2019-05-28 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Novel hyperthermophilic cellobiohydrolase II isolated from hot spring microbial community
To Be Published
|
|
7YCA
 
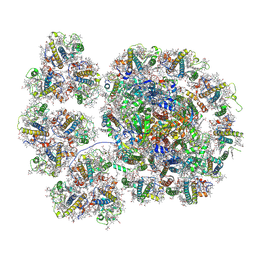 | | Cryo-EM structure of the PSI-LHCI-Lhcp supercomplex from Ostreococcus tauri | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (1~{S})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaenyl]cyclohex-3-en-1-ol, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Shan, J, Sheng, X, Ishii, A, Watanabe, A, Song, C, Murata, K, Minagawa, J, Liu, Z. | | Deposit date: | 2022-07-01 | | Release date: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | The photosystem I supercomplex from a primordial green alga Ostreococcus tauri harbors three light-harvesting complex trimers.
Elife, 12, 2023
|
|
8E40
 
 | | Full-length APOBEC3G in complex with HIV-1 Vif, CBF-beta, and fork RNA | | Descriptor: | Core-binding factor subunit beta, DNA dC->dU-editing enzyme APOBEC-3G, RNA, ... | | Authors: | Ito, F, Alvarez-Cabrera, A.L, Liu, S, Yang, H, Shiriaeva, A, Zhou, Z.H, Chen, X.S. | | Deposit date: | 2022-08-17 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structural basis for HIV-1 antagonism of host APOBEC3G via Cullin E3 ligase.
Sci Adv, 9, 2023
|
|
5DBE
 
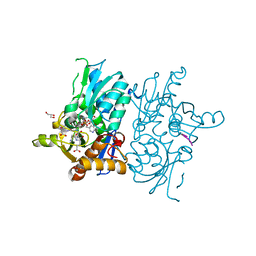 | | Crystal structure of O-acetylserine sulfhydrylase from Haemophilus influenzae in complex with pre-reactive O-acetyl serine, alpha-aminoacrylate reaction intermediate and peptide inhibitor at the resolution of 2.25A | | Descriptor: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, C-terminal peptide from Serine acetyltransferase, Cysteine synthase, ... | | Authors: | Singh, A.K, Kaushik, A, Ekka, M.K, Kumaran, S. | | Deposit date: | 2015-08-21 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure Of O-Acetylserine Sulfhydrylase From Haemophilus Influenzae In Complex With Pre-Reactive O-Acetyl Serine, Alpha-Aminoacrylate Reactionintermediate And Peptide Inhibitor At The Resolution Of 2.25A
To Be Published
|
|
7YA8
 
 | | The crystal structure of IpaH2.5 LRR domain | | Descriptor: | RING-type E3 ubiquitin transferase | | Authors: | Hiragi, K, Nishide, A, Takagi, K, Iwai, K, Kim, M, Mizushima, T. | | Deposit date: | 2022-06-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural insight into the recognition of the linear ubiquitin assembly complex by Shigella E3 ligase IpaH1.4/2.5.
J.Biochem., 173, 2023
|
|
7Y13
 
 | | Cryo-EM structure of apo-state MrgD-Gi complex (local) | | Descriptor: | PALMITIC ACID, Soluble cytochrome b562,Mas-related G-protein coupled receptor member D | | Authors: | Suzuki, S, Iida, M, Kawamoto, A, Oshima, A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-20 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into the activation mechanism of MrgD with heterotrimeric Gi-protein revealed by cryo-EM.
Commun Biol, 5, 2022
|
|
7Y12
 
 | | Cryo-EM structure of MrgD-Gi complex with beta-alanine | | Descriptor: | BETA-ALANINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Suzuki, S, Iida, M, Kawamoto, A, Oshima, A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-20 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into the activation mechanism of MrgD with heterotrimeric Gi-protein revealed by cryo-EM.
Commun Biol, 5, 2022
|
|
7Y14
 
 | | Cryo-EM structure of MrgD-Gi complex with beta-alanine (local) | | Descriptor: | BETA-ALANINE, PALMITIC ACID, Soluble cytochrome b562,Mas-related G-protein coupled receptor member D | | Authors: | Suzuki, S, Iida, M, Kawamoto, A, Oshima, A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-20 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insight into the activation mechanism of MrgD with heterotrimeric Gi-protein revealed by cryo-EM.
Commun Biol, 5, 2022
|
|
7Y15
 
 | | Cryo-EM structure of apo-state MrgD-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Suzuki, S, Iida, M, Kawamoto, A, Oshima, A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-20 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insight into the activation mechanism of MrgD with heterotrimeric Gi-protein revealed by cryo-EM.
Commun Biol, 5, 2022
|
|
8DZD
 
 | |
