5WVO
 
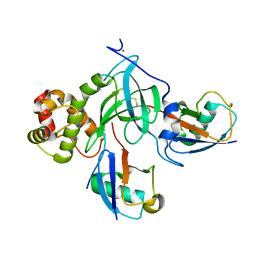 | | Crystal structure of DNMT1 RFTS domain in complex with K18/K23 mono-ubiquitylated histone H3 | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, Histone H3.1, Ubiquitin, ... | | Authors: | Ishiyama, S, Nishiyama, A, Nakanishi, M, Arita, K. | | Deposit date: | 2016-12-28 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structure of the Dnmt1 Reader Module Complexed with a Unique Two-Mono-Ubiquitin Mark on Histone H3 Reveals the Basis for DNA Methylation Maintenance
Mol. Cell, 68, 2017
|
|
5OL8
 
 | | Structure of human mitochondrial transcription elongation factor (TEFM) C-terminal domain | | Descriptor: | GLYCEROL, Transcription elongation factor, mitochondrial | | Authors: | Hillen, H.S, Parshin, A.V, Agaronyan, K, Morozov, Y, Graber, J.J, Chernev, A, Schwinghammer, K, Urlaub, H, Anikin, M, Cramer, P, Temiakov, D. | | Deposit date: | 2017-07-27 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of Transcription Anti-termination in Human Mitochondria.
Cell, 171, 2017
|
|
5X5I
 
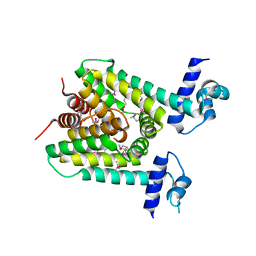 | | The X-ray crystal structure of a TetR family transcription regulator RcdA involved in the regulation of biofilm formation in Escherichia coli | | Descriptor: | HTH-type transcriptional regulator RcdA | | Authors: | Sugino, H, Usui, M, Shimada, T, Nakano, M, Ogasawara, H, Ishihama, A, Hirata, A. | | Deposit date: | 2017-02-16 | | Release date: | 2017-12-27 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | A structural sketch of RcdA, a transcription factor controlling the master regulator of biofilm formation.
FEBS Lett., 591, 2017
|
|
5ZFS
 
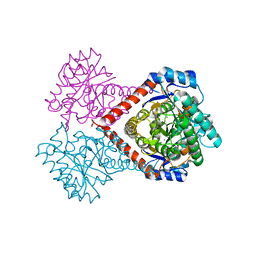 | | Crystal structure of Arthrobacter globiformis M30 sugar epimerase which can produce D-allulose from D-fructose | | Descriptor: | ACETATE ION, D-allulose-3-epimerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Yoshihara, A, Gullapalli, P.K, Ohtani, K, Akimitsu, K, Izumori, K, Kamitori, S. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | X-ray structure of Arthrobacter globiformis M30 ketose 3-epimerase for the production of D-allulose from D-fructose.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5OLA
 
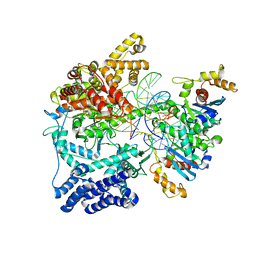 | | Structure of mitochondrial transcription elongation complex in complex with elongation factor TEFM | | Descriptor: | DNA (30-MER), DNA (5'-D(P*AP*TP*GP*GP*TP*GP*TP*AP*AP*CP*GP*CP*CP*AP*GP*AP*CP*GP*AP*AP*C)-3'), DNA-directed RNA polymerase, ... | | Authors: | Hillen, H.S, Parshin, A.V, Agaronyan, K, Morozov, Y, Graber, J.J, Chernev, A, Schwinghammer, K, Urlaub, H, Anikin, M, Cramer, P, Temiakov, D. | | Deposit date: | 2017-07-27 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.904 Å) | | Cite: | Mechanism of Transcription Anti-termination in Human Mitochondria.
Cell, 171, 2017
|
|
5OL9
 
 | | Structure of human mitochondrial transcription elongation factor (TEFM) N-terminal domain | | Descriptor: | ACETATE ION, Transcription elongation factor, mitochondrial | | Authors: | Hillen, H.S, Parshin, A.V, Agaronyan, K, Morozov, Y, Graber, J.J, Chernev, A, Schwinghammer, K, Urlaub, H, Anikin, M, Cramer, P, Temiakov, D. | | Deposit date: | 2017-07-27 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.302 Å) | | Cite: | Mechanism of Transcription Anti-termination in Human Mitochondria.
Cell, 171, 2017
|
|
5ODW
 
 | | Structure of the FpvAI-pyocin S2 complex | | Descriptor: | Ferripyoverdine receptor, Pyocin-S2, SULFATE ION | | Authors: | White, P, Joshi, A, Kleanthous, C. | | Deposit date: | 2017-07-06 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Exploitation of an iron transporter for bacterial protein antibiotic import.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XOY
 
 | | Crystal structure of LysK from Thermus thermophilus in complex with Lysine | | Descriptor: | LYSINE, SULFATE ION, [LysW]-lysine hydrolase | | Authors: | Tomita, T, Fujita, S, Hasebe, F, Cho, S.-H, Yoshida, A, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2017-05-31 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of LysK, an enzyme catalyzing the last step of lysine biosynthesis in Thermus thermophilus, in complex with lysine: Insight into the mechanism for recognition of the amino-group carrier protein, LysW
Biochem. Biophys. Res. Commun., 491, 2017
|
|
2D1X
 
 | | The crystal structure of the cortactin-SH3 domain and AMAP1-peptide complex | | Descriptor: | SULFATE ION, cortactin isoform a, proline rich region from development and differentiation enhancing factor 1 | | Authors: | Hashimoto, S, Hirose, M, Hashimoto, A, Morishige, M, Yamada, A, Hosaka, H, Akagi, K, Ogawa, E, Oneyama, C, Agatsuma, T, Okada, M, Kobayashi, H, Wada, H, Nakano, H, Ikegami, T, Nakagawa, A, Sabe, H. | | Deposit date: | 2005-09-01 | | Release date: | 2006-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting AMAP1 and cortactin binding bearing an atypical src homology 3/proline interface for prevention of breast cancer invasion and metastasis.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1X2R
 
 | | Structural basis for the defects of human lung cancer somatic mutations in the repression activity of Keap1 on Nrf2 | | Descriptor: | Kelch-like ECH-associated protein 1, Nuclear factor erythroid 2 related factor 2, SULFATE ION | | Authors: | Padmanabhan, B, Tong, K.I, Nakamura, Y, Ohta, T, Scharlock, M, Kobayashi, A, Ohtsuji, M, Kang, M.-I, Yamamoto, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-04-26 | | Release date: | 2006-03-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for defects of keap1 activity provoked by its point mutations in lung cancer
Mol.Cell, 21, 2006
|
|
2NCY
 
 | | Solution structure of pseudin-2 analog (Ps-P) | | Descriptor: | Pseudin-2 | | Authors: | Jeon, D, Kim, J, Shin, A, Kim, Y. | | Deposit date: | 2016-04-18 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Optimum Balance between the Cationicity and Structural Component for Bacterial Cell Selectivity and Anti-inflammatory activities of Pseudin-2 and its Analogs
To be Published
|
|
2CZL
 
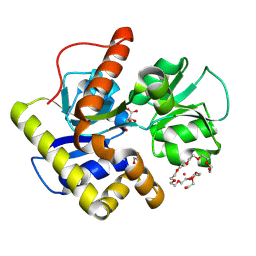 | | Crystal structure of MqnD (TTHA1568), a menaquinone biosynthetic enzyme from Thermus thermophilus HB8 (Cys11 modified with beta-mercaptoethanol) | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, L(+)-TARTARIC ACID, POTASSIUM ION, ... | | Authors: | Arai, R, Nishino, A, Nagano, K, Kamo-Uchikubo, T, Nishimoto, M, Toyama, M, Terada, T, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of MqnD (TTHA1568), a menaquinone biosynthetic enzyme from Thermus thermophilus HB8.
J.Struct.Biol., 168, 2009
|
|
2E0I
 
 | | Crystal structure of archaeal photolyase from Sulfolobus tokodaii with two FAD molecules: Implication of a novel light-harvesting cofactor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 432aa long hypothetical deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fujihashi, M, Numoto, N, Kobayashi, Y, Mizushima, A, Tsujimura, M, Nakamura, A, Kawarabayashi, Y, Miki, K. | | Deposit date: | 2006-10-10 | | Release date: | 2006-11-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Archaeal Photolyase from Sulfolobus tokodaii with Two FAD Molecules: Implication of a Novel Light-harvesting Cofactor
J.Mol.Biol., 365, 2007
|
|
2NCX
 
 | |
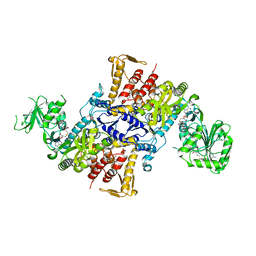
5X6Y
 
 | |
1X2J
 
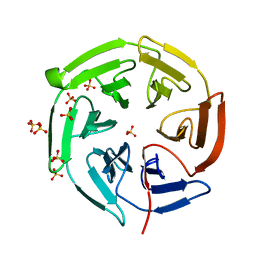 | | Structural basis for the defects of human lung cancer somatic mutations in the repression activity of Keap1 on Nrf2 | | Descriptor: | Kelch-like ECH-associated protein 1, SULFATE ION | | Authors: | Padmanabhan, B, Tong, K.I, Nakamura, Y, Ohta, T, Scharlock, M, Kobayashi, A, Ohtsuji, M, Kang, M.-I, Yamamoto, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-04-25 | | Release date: | 2006-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for defects of keap1 activity provoked by its point mutations in lung cancer
Mol.Cell, 21, 2006
|
|
2DUK
 
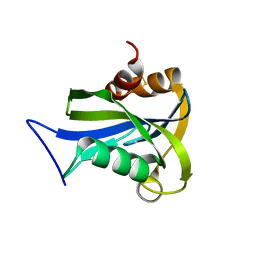 | | Crystal structure of MS0616 | | Descriptor: | MS0616 | | Authors: | Hosaka, T, Nishino, A, Uchikubo, K.-T, Kishishita, S, Murayama, K, Shirouzu, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-07-24 | | Release date: | 2007-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structure of MS0616
To be Published
|
|
5ZFG
 
 | | Crystal structure of a diazinon-metabolizing glutathione S-transferase in the silkworm, Bombyx mori | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Glutathione S-transferase | | Authors: | Yamamoto, K, Higashiura, A, Nakagawa, A. | | Deposit date: | 2018-03-06 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterisation of a diazinon-metabolising glutathione S-transferase in the silkworm Bombyx mori by X-ray crystallography and genome editing analysis.
Sci Rep, 8, 2018
|
|
5ZF1
 
 | | Molecular structure of a novel 5,10-methylenetetrahydrofolate dehydrogenase from the silkworm, Bombyx mori | | Descriptor: | 1,2-ETHANEDIOL, 5,10-methylenetetrahydrofolate dehydrogenase, SULFATE ION | | Authors: | Haque, R, Higashiura, A, Nakagawa, A, Yamamoto, K. | | Deposit date: | 2018-03-02 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular structure of a 5,10-methylenetetrahydrofolate dehydrogenase from the silkwormBombyx mori.
FEBS Open Bio, 9, 2019
|
|
2CWN
 
 | | Crystal structure of mouse transaldolase | | Descriptor: | Transaldolase | | Authors: | Handa, N, Arai, R, Nishino, A, Uchikubo, T, Takemoto, C, Morita, S, Kinoshita, Y, Nagano, Y, Uda, H, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-22 | | Release date: | 2005-12-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of mouse transaldolase
To be Published
|
|
5X6Z
 
 | |
1UG9
 
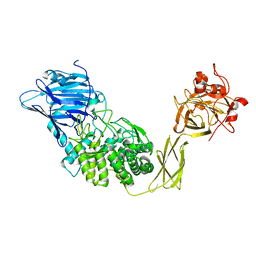 | | Crystal Structure of Glucodextranase from Arthrobacter globiformis I42 | | Descriptor: | CALCIUM ION, GLYCEROL, glucodextranase | | Authors: | Mizuno, M, Tonozuka, T, Suzuki, S, Uotsu-Tomita, R, Ohtaki, A, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2003-06-16 | | Release date: | 2003-12-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into substrate specificity and function of glucodextranase
J.Biol.Chem., 279, 2004
|
|
5Y9T
 
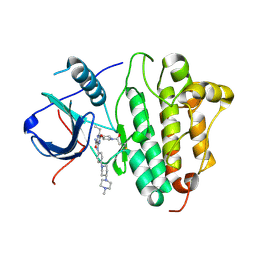 | | Crystal Structure of EGFR T790M mutant in complex with naquotinib | | Descriptor: | 6-ethyl-3-[[4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]amino]-5-[(3R)-1-prop-2-enoylpyrrolidin-3-yl]oxy-pyrazin e-2-carboxamide, Epidermal growth factor receptor | | Authors: | Mimasu, S, Tomimoto, Y, Maiko, I, Yasushi, A, Tatsuya, N. | | Deposit date: | 2017-08-28 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Pharmacological and Structural Characterizations of Naquotinib, a Novel Third-Generation EGFR Tyrosine Kinase Inhibitor, inEGFR-Mutated Non-Small Cell Lung Cancer.
Mol. Cancer Ther., 17, 2018
|
|
2ZJU
 
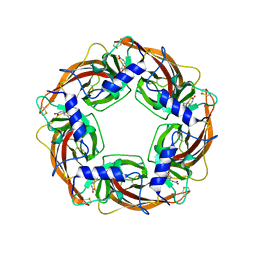 | | Crystal Structure of Lymnaea stagnalis Acetylcholine Binding Protein (Ls-AChBP) Complexed with Imidacloprid | | Descriptor: | (2E)-1-[(6-chloropyridin-3-yl)methyl]-N-nitroimidazolidin-2-imine, Acetylcholine-binding protein | | Authors: | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Morimoto, T, Matsuda, K. | | Deposit date: | 2008-03-10 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structures of Lymnaea stagnalis AChBP in complex with neonicotinoid insecticides imidacloprid and clothianidin
Invert.Neurosci., 8, 2008
|
|
1VB9
 
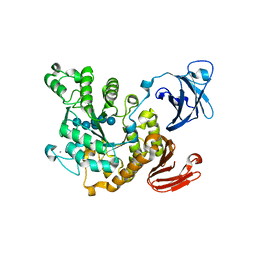 | | Crystal structure of Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) complexed with transglycosylated product | | Descriptor: | CALCIUM ION, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-[alpha-D-glucopyranose-(1-6)]alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-amylase II | | Authors: | Mizuno, M, Tonozuka, T, Uechi, A, Ohtaki, A, Ichikawa, K, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2004-02-25 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) complexed with transglycosylated product
EUR.J.BIOCHEM., 271, 2004
|
|
