5YCE
 
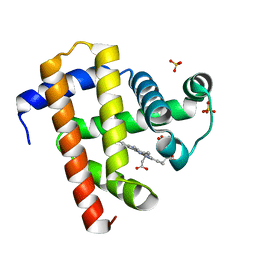 | | Sperm whale myoglobin swMb | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Isogai, Y, Imamura, H, Nakae, S, Sumi, T, Takahashi, K, Nakagawa, T, Tsuneshige, A, Shirai, T. | | Deposit date: | 2017-09-07 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.77 Å) | | Cite: | Tracing whale myoglobin evolution by resurrecting ancient proteins.
Sci Rep, 8, 2018
|
|
7KJZ
 
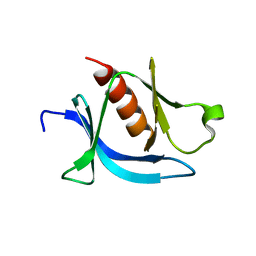 | | crystal structure of PLEKHA7 PH domain biding inositol-tetraphosphate | | Descriptor: | 1,2-ETHANEDIOL, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, Pleckstrin homology domain-containing family A member 7 | | Authors: | Marassi, F.M, Aleshin, A.E, Liddington, R.C. | | Deposit date: | 2020-10-26 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural basis for the association of PLEKHA7 with membrane-embedded phosphatidylinositol lipids.
Structure, 29, 2021
|
|
7KK7
 
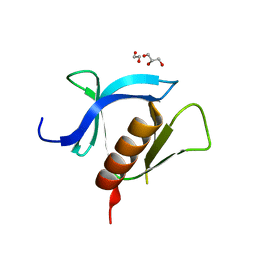 | | crystal structure of ligand-free PLEKHA7 PH domain | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Pleckstrin homology domain-containing family A member 7 | | Authors: | Marassi, F.M, Aleshin, A.E, Liddington, R.C. | | Deposit date: | 2020-10-27 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the association of PLEKHA7 with membrane-embedded phosphatidylinositol lipids.
Structure, 29, 2021
|
|
7KJO
 
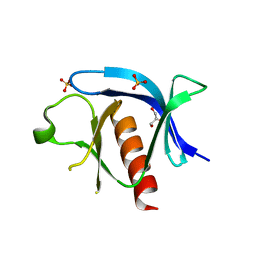 | |
5YCJ
 
 | | Ancestral myoglobin aMbWb' of Basilosaurus relative (polyphyly) imidazole-ligand | | Descriptor: | Ancestral myoglobin aMbWb' of Basilosaurus relative (polyphyly), IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Isogai, Y, Imamura, H, Nakae, S, Sumi, T, Takahashi, K, Nakagawa, T, Tsuneshige, A, Shirai, T. | | Deposit date: | 2017-09-07 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Tracing whale myoglobin evolution by resurrecting ancient proteins.
Sci Rep, 8, 2018
|
|
4LDS
 
 | |
1G59
 
 | | GLUTAMYL-TRNA SYNTHETASE COMPLEXED WITH TRNA(GLU). | | Descriptor: | GLUTAMYL-TRNA SYNTHETASE, TRNA(GLU) | | Authors: | Sekine, S, Nureki, O, Shimada, A, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-10-31 | | Release date: | 2001-09-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for anticodon recognition by discriminating glutamyl-tRNA synthetase.
Nat.Struct.Biol., 8, 2001
|
|
2H9U
 
 | | Crystal structure of the archaea specific DNA binding protein | | Descriptor: | 1,2-ETHANEDIOL, DNA/RNA-binding protein Alba 2 | | Authors: | Kumarevel, T.S, Sakamoto, K, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-06-12 | | Release date: | 2007-06-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the archaea specific DNA binding protein from Aeropyrum pernix K1
To be Published
|
|
7XKH
 
 | | Nucleotide-depleted F1 domain of FoF1-ATPase from Bacillus PS3, state1 | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit alpha, ... | | Authors: | Nakano, A, Kishikawa, J, Nakanishi, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-04-19 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of unisite catalysis of bacterial F 0 F 1 -ATPase.
Pnas Nexus, 1, 2022
|
|
7XKO
 
 | | F1 domain of epsilon C-terminal domain deleted FoF1 from Bacillus PS3,state1,nucleotide depeleted | | Descriptor: | ATP synthase gamma chain, ATP synthase subunit alpha, ATP synthase subunit beta, ... | | Authors: | Nakano, A, Kishikawa, J, Nakanishi, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-04-20 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of unisite catalysis of bacterial F 0 F 1 -ATPase.
Pnas Nexus, 1, 2022
|
|
7XKQ
 
 | | F1 domain of FoF1-ATPase with the down form of epsilon subunit from Bacillus PS3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Nakano, A, Kishikawa, J, Nakanishi, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-04-20 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of unisite catalysis of bacterial F 0 F 1 -ATPase.
Pnas Nexus, 1, 2022
|
|
7XKR
 
 | | F1 domain of FoF1-ATPase with the up form of epsilon subunit from Bacillus PS3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Nakano, A, Kishikawa, J, Nakanishi, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-04-20 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of unisite catalysis of bacterial F 0 F 1 -ATPase.
Pnas Nexus, 1, 2022
|
|
7XKP
 
 | | F1 domain of epsilon C-terminal domain deleted FoF1 from Bacillus PS3,state1,unisite condition | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase gamma chain, ATP synthase subunit alpha, ... | | Authors: | Nakano, A, Kishikawa, J, Nakanishi, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-04-20 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of unisite catalysis of bacterial F 0 F 1 -ATPase.
Pnas Nexus, 1, 2022
|
|
2GZW
 
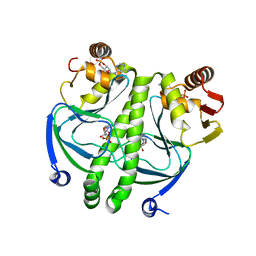 | | Crystal structure of the E.coli CRP-cAMP complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator | | Authors: | Kumarevel, T.S, Tanaka, T, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-05-12 | | Release date: | 2007-05-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of activated CRP protein from E coli
To be Published
|
|
3WVL
 
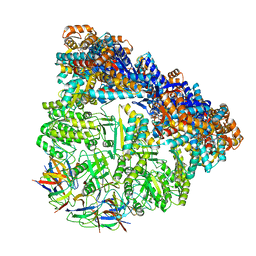 | | Crystal structure of the football-shaped GroEL-GroES complex (GroEL: GroES2:ATP14) from Escherichia coli | | Descriptor: | 10 kDa chaperonin, 60 kDa chaperonin, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Koike-Takeshita, A, Arakawa, T, Taguchi, H, Shimamura, T. | | Deposit date: | 2014-05-23 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.788 Å) | | Cite: | Crystal structure of a symmetric football-shaped GroEL:GroES2-ATP14 complex determined at 3.8 angstrom reveals rearrangement between two GroEL rings.
J.Mol.Biol., 426, 2014
|
|
7F81
 
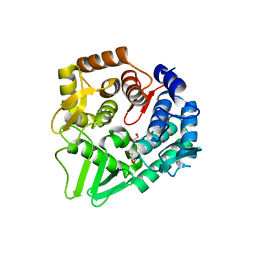 | | Structure of the bacterial cellulose synthase subunit Z from Enterobacter sp. CJF-002 | | Descriptor: | GLYCEROL, Glucanase, S,R MESO-TARTARIC ACID | | Authors: | Fujiwara, T, Fujishima, A, Yao, M. | | Deposit date: | 2021-06-30 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural snapshot of a glycoside hydrolase family 8 endo-beta-1,4-glucanase capturing the state after cleavage of the scissile bond.
Acta Crystallogr.,Sect.D, 78, 2022
|
|
7F82
 
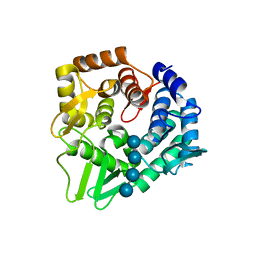 | | Structure of the bacterial cellulose synthase subunit Z in complex with cellooligosaccharides from Enterobacter sp. CJF-002 | | Descriptor: | Glucanase, S,R MESO-TARTARIC ACID, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Fujiwara, T, Fujishima, A, Yao, M. | | Deposit date: | 2021-06-30 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural snapshot of a glycoside hydrolase family 8 endo-beta-1,4-glucanase capturing the state after cleavage of the scissile bond.
Acta Crystallogr.,Sect.D, 78, 2022
|
|
3VU9
 
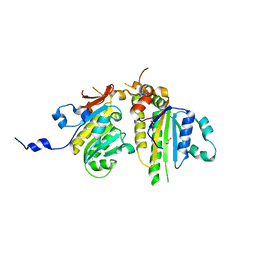 | | Crystal Structure of Psy3-Csm2 complex | | Descriptor: | 1,2-ETHANEDIOL, Chromosome segregation in meiosis protein 2, Platinum sensitivity protein 3 | | Authors: | Tawaramoto, M, Sasanuma, H, Hosaka, H, Lao, J.P, Sanda, E, Suzuki, M, Yamashita, E, Hunter, N, Shinohara, M, Nakagawa, A, Shinohara, A. | | Deposit date: | 2012-06-23 | | Release date: | 2013-04-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A new protein complex promoting the assembly of Rad51 filaments
Nat Commun, 4, 2013
|
|
6AIF
 
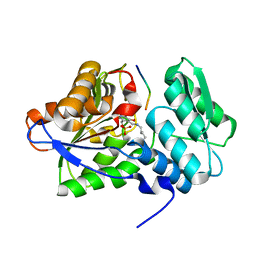 | |
1IO0
 
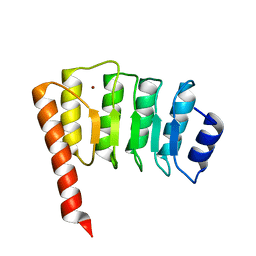 | | CRYSTAL STRUCTURE OF TROPOMODULIN C-TERMINAL HALF | | Descriptor: | TROPOMODULIN, ZINC ION | | Authors: | Krieger, I, Kostyukova, A, Yamashita, A, Maeda, Y. | | Deposit date: | 2000-12-14 | | Release date: | 2002-11-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of the C-terminal half of tropomodulin and structural basis of actin filament pointed-end capping.
Biophys.J., 83, 2002
|
|
1EWZ
 
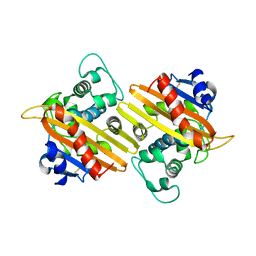 | | CRYSTAL STRUCTURE OF THE OXA-10 BETA-LACTAMASE FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | BETA LACTAMASE OXA-10 | | Authors: | Golemi, D, Maveyraud, L, Vakulenko, S, Tranier, S, Ishiwata, A, Kotra, L.P, Samama, J.P, Mobashery, S. | | Deposit date: | 2000-04-28 | | Release date: | 2000-11-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The First Structural and Mechanistic Insights for Class D beta-Lactamases: Evidence for a Novel Catalytic Process for Turnover of beta-Lactam Antibiotics
J.Am.Chem.Soc., 122, 2000
|
|
2SRC
 
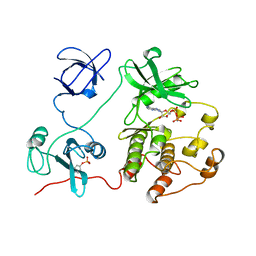 | | CRYSTAL STRUCTURE OF HUMAN TYROSINE-PROTEIN KINASE C-SRC, IN COMPLEX WITH AMP-PNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, TYROSINE-PROTEIN KINASE SRC | | Authors: | Xu, W, Doshi, A, Lei, M, Eck, M.J, Harrison, S.C. | | Deposit date: | 1998-12-29 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of c-Src reveal features of its autoinhibitory mechanism.
Mol.Cell, 3, 1999
|
|
7W64
 
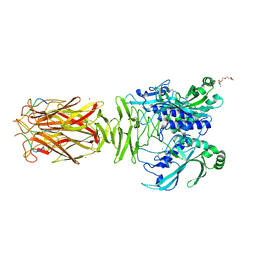 | | Crystal structure of minor pilin TcpB from Vibrio cholerae complexed with N-terminal peptide fragment of TcpF | | Descriptor: | CALCIUM ION, CHLORIDE ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Oki, H, Kawahara, K, Iimori, M, Imoto, Y, Maruno, T, Uchiyama, S, Muroga, Y, Yoshida, A, Yoshida, T, Ohkubo, T, Matsuda, S, Iida, T, Nakamura, S. | | Deposit date: | 2021-12-01 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the toxin-coregulated pilus-dependent secretion of Vibrio cholerae colonization factor.
Sci Adv, 8, 2022
|
|
7W65
 
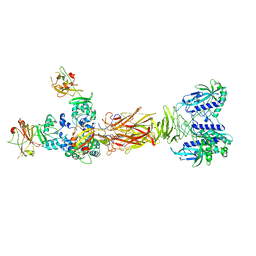 | | Crystal structure of minor pilin TcpB from Vibrio cholerae complexed with secreted protein TcpF | | Descriptor: | Toxin coregulated pilus biosynthesis protein F, Toxin-coregulated pilus biosynthesis protein B | | Authors: | Oki, H, Kawahara, K, Iimori, M, Imoto, Y, Maruno, T, Uchiyama, S, Muroga, Y, Yoshida, A, Yoshida, T, Ohkubo, T, Matsuda, S, Iida, T, Nakamura, S. | | Deposit date: | 2021-12-01 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.05 Å) | | Cite: | Structural basis for the toxin-coregulated pilus-dependent secretion of Vibrio cholerae colonization factor.
Sci Adv, 8, 2022
|
|
7W63
 
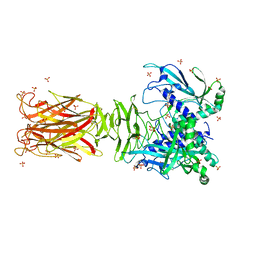 | | Crystal structure of minor pilin TcpB from Vibrio cholerae | | Descriptor: | SULFATE ION, Toxin-coregulated pilus biosynthesis protein B | | Authors: | Oki, H, Kawahara, K, Iimori, M, Imoto, Y, Maruno, T, Uchiyama, S, Muroga, Y, Yoshida, A, Yoshida, T, Ohkubo, T, Matsuda, S, Iida, T, Nakamura, S. | | Deposit date: | 2021-12-01 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-16 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural basis for the toxin-coregulated pilus-dependent secretion of Vibrio cholerae colonization factor.
Sci Adv, 8, 2022
|
|
