3WKZ
 
 | | Crystal Structure of the Ostrinia furnacalis Group I Chitinase catalytic domain E148Q mutant | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase | | 著者 | Chen, L, Liu, T, Zhou, Y, Chen, Q, Shen, X, Yang, Q. | | 登録日 | 2013-11-05 | | 公開日 | 2014-04-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Structural characteristics of an insect group I chitinase, an enzyme indispensable to moulting.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WL1
 
 | | Crystal structure of Ostrinia furnacalis Group I chitinase catalytic domain in complex with reaction products (GlcNAc)2,3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Chen, L, Liu, T, Zhou, Y, Chen, Q, Shen, X, Yang, Q. | | 登録日 | 2013-11-05 | | 公開日 | 2014-04-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.772 Å) | | 主引用文献 | Structural characteristics of an insect group I chitinase, an enzyme indispensable to moulting.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3R5M
 
 | | Crystal structure of RXRalphaLBD complexed with the agonist magnolol | | 分子名称: | 5,5'-di(prop-2-en-1-yl)biphenyl-2,2'-diol, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | 著者 | Zhang, H, Chen, L, Chen, J, Hu, L, Jiang, H, Shen, X. | | 登録日 | 2011-03-18 | | 公開日 | 2012-02-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Molecular determinants of magnolol targeting both RXR(alpha) and PPAR(gamma).
Plos One, 6, 2011
|
|
3R29
 
 | | Crystal structure of RXRalpha ligand-binding domain complexed with corepressor SMRT2 | | 分子名称: | Nuclear receptor corepressor 2, Retinoic acid receptor RXR-alpha | | 著者 | Zhang, H, Chen, L, Chen, J, Jiang, H, Shen, X. | | 登録日 | 2011-03-14 | | 公開日 | 2011-05-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis for retinoic x receptor repression on the tetramer.
J.Biol.Chem., 286, 2011
|
|
3R2A
 
 | | Crystal structure of RXRalpha ligand-binding domain complexed with corepressor SMRT2 and antagonist rhein | | 分子名称: | 4,5-dihydroxy-9,10-dioxo-9,10-dihydroanthracene-2-carboxylic acid, Nuclear receptor corepressor 2, Retinoic acid receptor RXR-alpha | | 著者 | Zhang, H, Chen, L, Chen, J, Jiang, H, Shen, X. | | 登録日 | 2011-03-14 | | 公開日 | 2011-05-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural basis for retinoic x receptor repression on the tetramer.
J.Biol.Chem., 286, 2011
|
|
3F9G
 
 | |
3F9F
 
 | |
3F9H
 
 | |
3F9E
 
 | |
3R5N
 
 | | Crystal structure of PPARgammaLBD complexed with the agonist magnolol | | 分子名称: | 5,5'-di(prop-2-en-1-yl)biphenyl-2,2'-diol, Peroxisome proliferator-activated receptor gamma | | 著者 | Zhang, H, Chen, L, Chen, J, Hu, L, Jiang, H, Shen, X. | | 登録日 | 2011-03-18 | | 公開日 | 2012-02-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Molecular determinants of magnolol targeting both RXR(alpha) and PPAR(gamma).
Plos One, 6, 2011
|
|
3FXJ
 
 | | Crystal Structure of Human Protein phosphatase 1A (PPM1A) Bound with Phosphate at 3 mM of Mn2+ | | 分子名称: | MANGANESE (II) ION, PHOSPHATE ION, Protein phosphatase 1A | | 著者 | Hu, T, Wang, L, Wang, K, Jiang, H, Shen, X. | | 登録日 | 2009-01-21 | | 公開日 | 2010-01-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for the Mn2+-dependent activation of human PPM1A
To be published
|
|
3FXK
 
 | | Crystal Structure of Human Protein phosphatase 1A (PPM1A) Bound with Phosphate at 10 mM of Mn2+ | | 分子名称: | MANGANESE (II) ION, PHOSPHATE ION, Protein phosphatase 1A | | 著者 | Hu, T, Wang, L, Wang, K, Jiang, H, Shen, X. | | 登録日 | 2009-01-21 | | 公開日 | 2010-01-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for the Mn2+-dependent activation of human PPM1A
To be published
|
|
3FXL
 
 | | Crystal Structure of Human Protein phosphatase 1A (PPM1A) Bound with Citrate at 1 mM of Mn2+ | | 分子名称: | CITRATE ANION, MANGANESE (II) ION, PHOSPHATE ION, ... | | 著者 | Hu, T, Wang, L, Wang, K, Jiang, H, Shen, X. | | 登録日 | 2009-01-21 | | 公開日 | 2010-01-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for the Mn2+-dependent activation of human PPM1A
To be published
|
|
3FXO
 
 | | Crystal Structure of Human Protein phosphatase 1A (PPM1A) Bound with Phosphate at 1 mM of Mn2+ | | 分子名称: | MANGANESE (II) ION, PHOSPHATE ION, Protein phosphatase 1A | | 著者 | Hu, T, Wang, L, Wang, K, Jiang, H, Shen, X. | | 登録日 | 2009-01-21 | | 公開日 | 2010-01-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for the Mn2+-dependent activation of human PPM1A
To be published
|
|
3FXM
 
 | | Crystal Structure of Human Protein phosphatase 1A (PPM1A) Bound with Citrate at 10 mM of Mn2+ | | 分子名称: | CITRATE ANION, MANGANESE (II) ION, PHOSPHATE ION, ... | | 著者 | Hu, T, Wang, L, Wang, K, Jiang, H, Shen, X. | | 登録日 | 2009-01-21 | | 公開日 | 2010-01-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for the Mn2+-dependent activation of human PPM1A
To be published
|
|
3OZO
 
 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with NGT | | 分子名称: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, N-acetylglucosaminidase | | 著者 | Zhang, H, Liu, T, Liu, F, Shen, X, Yang, Q. | | 登録日 | 2010-09-27 | | 公開日 | 2011-09-28 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with NGT
To be Published
|
|
3OZP
 
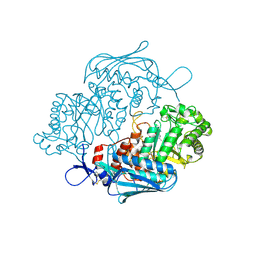 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with PUGNAc | | 分子名称: | N-acetylglucosaminidase, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE | | 著者 | Zhang, H, Liu, T, Liu, F, Shen, X, Yang, Q. | | 登録日 | 2010-09-27 | | 公開日 | 2011-08-17 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Active-pocket size differentiating insectile from bacterial chitinolytic beta-N-acetyl-D-hexosaminidases
Biochem.J., 438, 2011
|
|
2RJG
 
 | | Crystal structure of biosynthetic alaine racemase from Escherichia coli | | 分子名称: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | 登録日 | 2007-10-15 | | 公開日 | 2008-07-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
3NSN
 
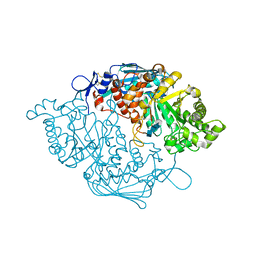 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with TMG-chitotriomycin | | 分子名称: | 2-deoxy-2-(trimethylammonio)-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetylglucosaminidase | | 著者 | Zhang, H, Liu, T, Liu, F, Yang, Q, Shen, X. | | 登録日 | 2010-07-02 | | 公開日 | 2010-11-24 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Determinants of an Insect {beta}-N-Acetyl-D-hexosaminidase Specialized as a Chitinolytic Enzyme
J.Biol.Chem., 286, 2011
|
|
3NSM
 
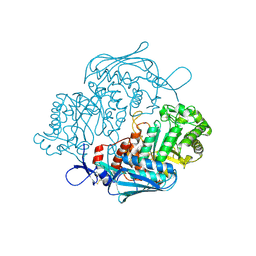 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 from Ostrinia furnacalis | | 分子名称: | N-acetylglucosaminidase | | 著者 | Zhang, H, Liu, T, Liu, F, Yang, Q, Shen, X. | | 登録日 | 2010-07-02 | | 公開日 | 2010-11-24 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Determinants of an Insect {beta}-N-Acetyl-D-hexosaminidase Specialized as a Chitinolytic Enzyme
J.Biol.Chem., 286, 2011
|
|
2QGH
 
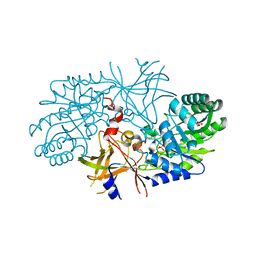 | | Crystal structure of diaminopimelate decarboxylase from Helicobacter pylori complexed with L-lysine | | 分子名称: | Diaminopimelate decarboxylase, GLYCEROL, LYSINE, ... | | 著者 | Hu, T, Wu, D, Jiang, H, Shen, X. | | 登録日 | 2007-06-28 | | 公開日 | 2008-05-27 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of diaminopimelate decarboxylase from Helicobacter pylori complexed with L-lysine
TO BE PUBLISHED
|
|
3S6T
 
 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 V327G complexed with PUGNAc | | 分子名称: | N-acetylglucosaminidase, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE | | 著者 | Zhang, H, Liu, T, Yang, Q, Shen, X. | | 登録日 | 2011-05-26 | | 公開日 | 2012-04-11 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Active-pocket size differentiating insectile from bacterial chitinolytic beta-N-acetyl-D-hexosaminidases.
Biochem.J., 438, 2011
|
|
2PWX
 
 | |
2RJH
 
 | | Crystal structure of biosynthetic alaine racemase in D-cycloserine-bound form from Escherichia coli | | 分子名称: | Alanine racemase, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, SULFATE ION | | 著者 | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | 登録日 | 2007-10-15 | | 公開日 | 2008-07-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
7DKL
 
 | |
