7D1T
 
 | | Cryo-EM Structure of PSII at 1.95 angstrom resolution | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Kato, K, Miyazaki, N, Hamaguchi, T, Nakajima, Y, Akita, F, Yonekura, K, Shen, J.R. | | Deposit date: | 2020-09-15 | | Release date: | 2021-03-31 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (1.95 Å) | | Cite: | High-resolution cryo-EM structure of photosystem II reveals damage from high-dose electron beams.
Commun Biol, 4, 2021
|
|
7D1U
 
 | | Cryo-EM Structure of PSII at 2.08 angstrom resolution | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Kato, K, Miyazaki, N, Hamaguchi, T, Nakajima, Y, Akita, F, Yonekura, K, Shen, J.R. | | Deposit date: | 2020-09-15 | | Release date: | 2021-03-31 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (2.08 Å) | | Cite: | High-resolution cryo-EM structure of photosystem II reveals damage from high-dose electron beams.
Commun Biol, 4, 2021
|
|
7DZI
 
 | | intermediate of FABP with a delay time of 300 ns | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZH
 
 | | intermediate of FABP with a delay time of 100 ns | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZJ
 
 | | Fabp protein before hv | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZK
 
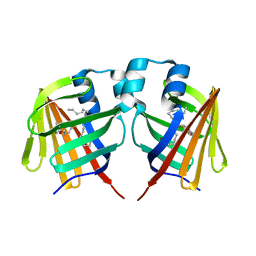 | | Fabp protein after hv | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZL
 
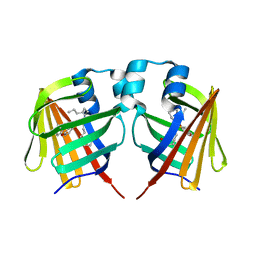 | | A69C-M71L mutant of Fabp protein | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
4XK8
 
 | | Crystal structure of plant photosystem I-LHCI super-complex at 2.8 angstrom resolution | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Suga, M, Qin, X, Kuang, T, Shen, J.R. | | Deposit date: | 2015-01-10 | | Release date: | 2015-06-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for energy transfer pathways in the plant PSI-LHCI supercomplex
Science, 348, 2015
|
|
7EW6
 
 | | Barley photosystem I-LHCI-Lhca5 supercomplex | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Wang, W.D, Shen, L, Tang, K, Han, G.Y, Zhang, X, Shen, J.R. | | Deposit date: | 2021-05-24 | | Release date: | 2021-12-22 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Architecture of the chloroplast PSI-NDH supercomplex in Hordeum vulgare.
Nature, 601, 2022
|
|
7EU3
 
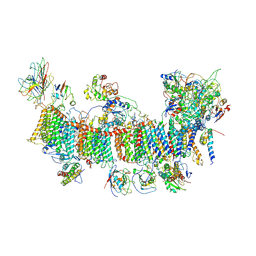 | | Chloroplast NDH complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, ... | | Authors: | Wang, W.D, Shen, L, Tang, K, Han, G.Y, Zhang, X, Shen, J.R. | | Deposit date: | 2021-05-15 | | Release date: | 2021-12-29 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Architecture of the chloroplast PSI-NDH supercomplex in Hordeum vulgare.
Nature, 601, 2022
|
|
7EWK
 
 | | Barley photosystem I-LHCI-Lhca6 supercomplex | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Wang, W.D, Shen, L, Tang, K, Han, G.Y, Zhang, X, Shen, J.R. | | Deposit date: | 2021-05-25 | | Release date: | 2022-01-12 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Architecture of the chloroplast PSI-NDH supercomplex in Hordeum vulgare.
Nature, 601, 2022
|
|
8AP1
 
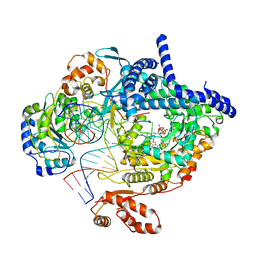 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with two GTP molecules poised for de novo initiation (IC2) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2022-08-09 | | Release date: | 2023-08-23 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8ATV
 
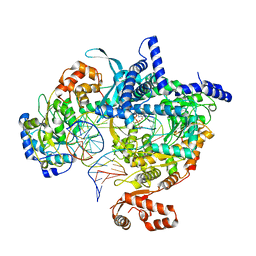 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 5-mer RNA, pppGpGpApApA (IC5) | | Descriptor: | DNA (36-MER), DNA-directed RNA polymerase, mitochondrial, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8ATW
 
 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 6-mer RNA, pppGpGpApApApU (IC6) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8ATT
 
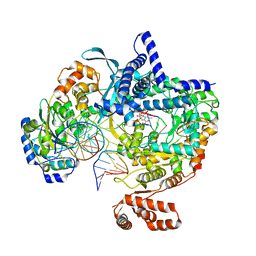 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 4-mer RNA, pppGpGpUpA (IC4) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
6L4T
 
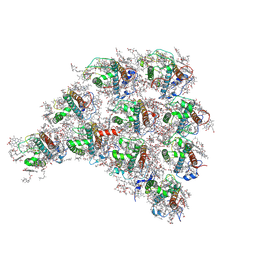 | | Structure of the peripheral FCPI from diatom | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nagao, R, Kato, K, Miyazaki, N, Akita, F, Shen, J.R. | | Deposit date: | 2019-10-21 | | Release date: | 2020-05-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of photosystem I-light-harvesting supercomplex from a red-lineage diatom
Nat Commun, 2020
|
|
4WM8
 
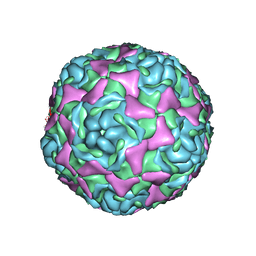 | | Crystal Structure of Human Enterovirus D68 | | Descriptor: | DECANOIC ACID, VP1, VP2, ... | | Authors: | Liu, Y, Sheng, J, Fokine, A, Meng, G, Long, F, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2014-10-08 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Virus structure. Structure and inhibition of EV-D68, a virus that causes respiratory illness in children.
Science, 347, 2015
|
|
6JEO
 
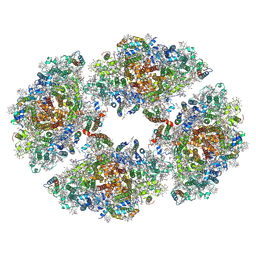 | | Structure of PSI tetramer from Anabaena | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Nagao, R, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-02-06 | | Release date: | 2019-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a cyanobacterial photosystem I tetramer revealed by cryo-electron microscopy.
Nat Commun, 10, 2019
|
|
8C5U
 
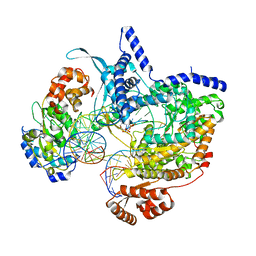 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 8-mer RNA, pppGpGpUpApApApUpG (IC8) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2023-01-10 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8C5S
 
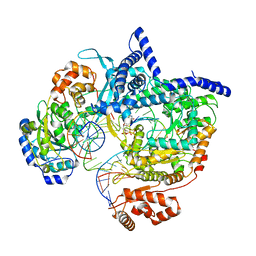 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 7-mer RNA, pppGpGpUpApApApU (IC7) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2023-01-10 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
6KMW
 
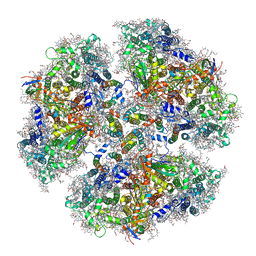 | | Structure of PSI from H. hongdechloris grown under white light condition | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Nagao, R, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-08-01 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Structural basis for the adaptation and function of chlorophyll f in photosystem I.
Nat Commun, 11, 2020
|
|
6KMX
 
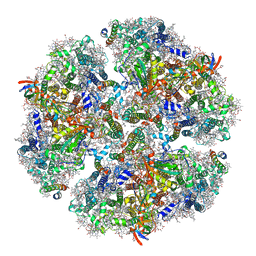 | | Structure of PSI from H. hongdechloris grown under far-red light condition | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Nagao, R, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-08-01 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Structural basis for the adaptation and function of chlorophyll f in photosystem I.
Nat Commun, 11, 2020
|
|
6TAS
 
 | | Structure of the two-fold capsomer of the dArc1 capsid | | Descriptor: | Activity-regulated cytoskeleton associated protein 1, ZINC ION | | Authors: | Erlendsson, S, Morado, D.R, Shepherd, J.D, Briggs, J.A.G. | | Deposit date: | 2019-10-30 | | Release date: | 2020-01-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structures of virus-like capsids formed by the Drosophila neuronal Arc proteins.
Nat.Neurosci., 23, 2020
|
|
6K33
 
 | | Structure of PSI-isiA supercomplex from Thermosynechococcus vulcanus | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Akita, F, Nagao, R, Kato, K, Shen, J.R, Miyazaki, N. | | Deposit date: | 2019-05-16 | | Release date: | 2020-05-20 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structure of a cyanobacterial photosystem I surrounded by octadecameric IsiA antenna proteins.
Commun Biol, 3, 2020
|
|
7DR1
 
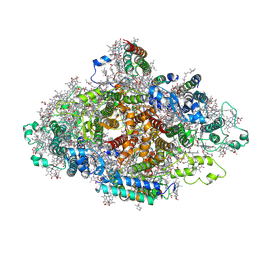 | | Structure of Wild-type PSI monomer2 from Cyanophora paradoxa | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Nagao, R, Akita, F, Miyazaki, N, Shen, J.R. | | Deposit date: | 2020-12-25 | | Release date: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into an evolutionary turning-point of photosystem I from prokaryotes to eukaryotes
Biorxiv, 2022
|
|
