2L0J
 
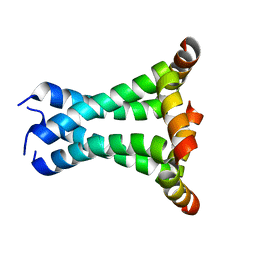 | | Solid State NMR structure of the M2 proton channel from Influenza A Virus in hydrated lipid bilayer | | Descriptor: | Matrix protein 2 | | Authors: | Sharma, M, Yi, M, Dong, H, Qin, H, Peterson, E, Busath, D.D, Zhou, H.X, Cross, T.A. | | Deposit date: | 2010-07-08 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Insight into the mechanism of the influenza a proton channel from a structure in a lipid bilayer.
Science, 330, 2010
|
|
4V61
 
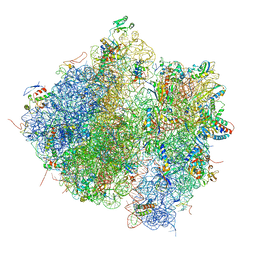 | | Homology model for the Spinach chloroplast 30S subunit fitted to 9.4A cryo-EM map of the 70S chlororibosome. | | Descriptor: | 16S rRNA, 23S rRNA, 4.8S rRNA, ... | | Authors: | Sharma, M.R, Wilson, D.N, Datta, P.P, Barat, C, Schluenzen, F, Fucini, P, Agrawal, R.K. | | Deposit date: | 2007-11-09 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Cryo-EM study of the spinach chloroplast ribosome reveals the structural and functional roles of plastid-specific ribosomal proteins
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7OLF
 
 | |
7OH2
 
 | |
7OFX
 
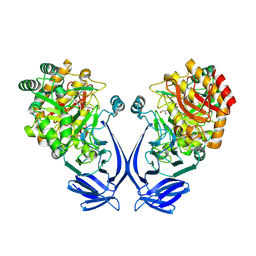 | | Crystal structure of a GH31 family sulfoquinovosidase mutant D455N from Agrobacterium tumefaciens in complex with sulfoquinovosyl glycerol (SQGro) | | Descriptor: | Alpha-glucosidase yihQ, [(2S,3S,4S,5R,6S)-6-[(2R)-2,3-bis(oxidanyl)propoxy]-3,4,5-tris(oxidanyl)oxan-2-yl]methanesulfonic acid | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2021-05-05 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Oxidative desulfurization pathway for complete catabolism of sulfoquinovose by bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7NE2
 
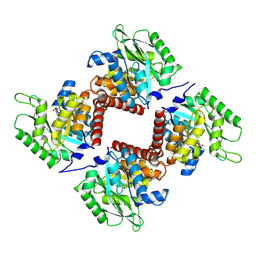 | | Crystal structure of class I SFP aldolase YihT from Salmonella enterica with SFP/ DHAP (Schiff base complex with active site Lys193) | | Descriptor: | (2~{S},3~{S},4~{R})-2,3,4,5-tetrakis(oxidanyl)-6-phosphonooxy-hexane-1-sulfonic acid, Sulfofructosephosphate aldolase, [(~{E})-2,3-bis(oxidanyl)prop-1-enyl] dihydrogen phosphate | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2021-02-03 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular Basis of Sulfosugar Selectivity in Sulfoglycolysis.
Acs Cent.Sci., 7, 2021
|
|
8C54
 
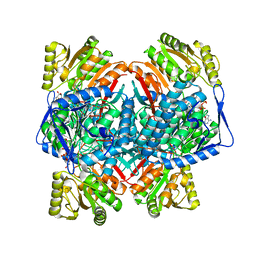 | | Cryo-EM structure of NADH bound SLA dehydrogenase RlGabD from Rhizobium leguminosarum bv. trifolii SRD1565 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Succinate semialdehyde dehydrogenase | | Authors: | Sharma, M, Meek, R.W, Armstrong, Z, Blaza, J.N, Alhifthi, A, Li, J, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2023-01-06 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Molecular basis of sulfolactate synthesis by sulfolactaldehyde dehydrogenase from Rhizobium leguminosarum.
Chem Sci, 14, 2023
|
|
7AGH
 
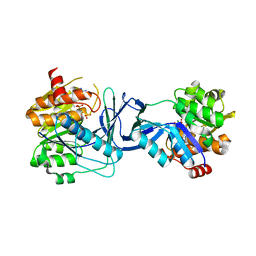 | |
7AG1
 
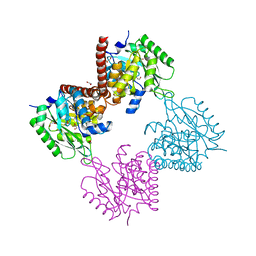 | |
8P2J
 
 | |
7AG4
 
 | |
7AG7
 
 | |
8Q57
 
 | | Crystal structure of class II SFP aldolase from Yersinia aldovae (YaSqiA-Zn-SO4) with bound sulfate ions | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, SULFATE ION, ... | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2023-08-08 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Defining the molecular architecture, metal dependence, and distribution of metal-dependent class II sulfofructose-1-phosphate aldolases.
J.Biol.Chem., 299, 2023
|
|
6NF8
 
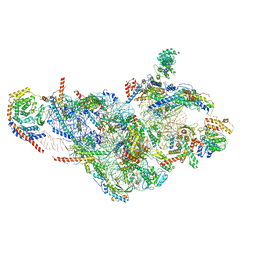 | | Structure of human mitochondrial translation initiation factor 3 bound to the small ribosomal subunit -Class I | | Descriptor: | 28S ribosomal RNA, mitochondria, 28S ribosomal protein S10, ... | | Authors: | Sharma, M, Koripella, R, Agrawal, R. | | Deposit date: | 2018-12-19 | | Release date: | 2019-02-27 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structure of Human Mitochondrial Translation Initiation Factor 3 Bound to the Small Ribosomal Subunit.
iScience, 12, 2019
|
|
6NEQ
 
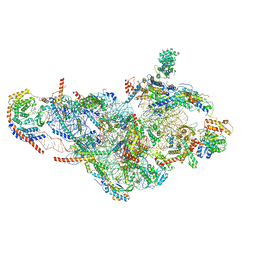 | | Structure of human mitochondrial translation initiation factor 3 bound to the small ribosomal subunit-Class-II | | Descriptor: | 28S ribosomal RNA, mitochondrial, 28S ribosomal protein S10, ... | | Authors: | Sharma, M, Koripella, R, Agrawal, R. | | Deposit date: | 2018-12-18 | | Release date: | 2019-02-27 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structure of Human Mitochondrial Translation Initiation Factor 3 Bound to the Small Ribosomal Subunit.
iScience, 12, 2019
|
|
8R56
 
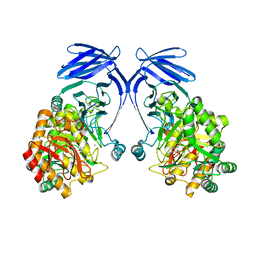 | |
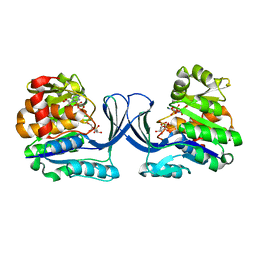
7AG6
 
 | |
7AGK
 
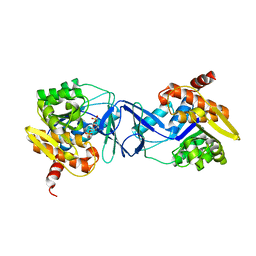 | | Crystal structure of E. coli SF kinase (YihV) in complex with product sulfofructose phosphate (SFP) | | Descriptor: | Sulfofructose kinase, [(2~{S},3~{S},4~{S},5~{R})-3,4,5-tris(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]methanesulfonic acid | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2020-09-22 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Molecular Basis of Sulfosugar Selectivity in Sulfoglycolysis.
Acs Cent.Sci., 7, 2021
|
|
8FMW
 
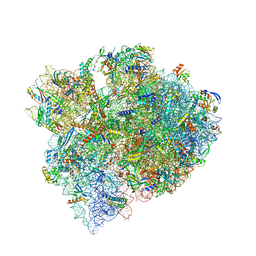 | | The structure of a hibernating ribosome in the Lyme disease pathogen | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Sharma, M.R, Manjari, S.R, Agrawal, E.K, Keshavan, P, Koripella, R.K, Majumdar, S, Marcinkiewicz, A.L, Lin, Y.P, Agrawal, R.K, Banavali, N.K. | | Deposit date: | 2022-12-25 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | The structure of a hibernating ribosome in a Lyme disease pathogen.
Nat Commun, 14, 2023
|
|
8FN2
 
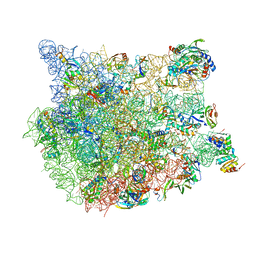 | | The structure of a 50S ribosomal subunit in the Lyme disease pathogen Borreliella burgdorferi | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Sharma, M.R, Manjari, S.R, Agrawal, E.K, Keshavan, P, Koripella, R.K, Majumdar, S, Marcinkiewicz, A.L, Lin, Y.P, Agrawal, R.K, Banavali, N.K. | | Deposit date: | 2022-12-26 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structure of a hibernating ribosome in a Lyme disease pathogen.
Nat Commun, 14, 2023
|
|
8Q59
 
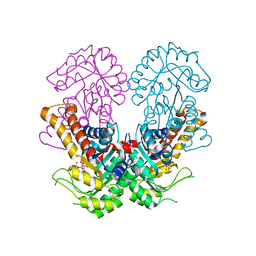 | | Crystal structure of metal-dependent class II sulfofructose phosphate aldolase from Yersinia aldovae in complex with sulfofructose phosphate (YaSqiA-Zn-SFP) | | Descriptor: | (3~{S},4~{S})-2,3,4-tris(oxidanyl)-5-oxidanylidene-6-phosphonooxy-hexane-1-sulfonic acid, SODIUM ION, Tagatose-1,6-bisphosphate aldolase kbaY, ... | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2023-08-08 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Defining the molecular architecture, metal dependence, and distribution of metal-dependent class II sulfofructose-1-phosphate aldolases.
J.Biol.Chem., 299, 2023
|
|
8QC5
 
 | |
6DZP
 
 | | Cryo-EM Structure of Mycobacterium smegmatis C(minus) 50S ribosomal subunit | | Descriptor: | 23S rRNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Sharma, M.R, Li, Y, Korripella, R, Yang, Y, Kaushal, P.S, Lin, Q, Wade, J.T, Gray, A.G, Derbyshire, K.M, Agrawal, R.K, Ojha, A. | | Deposit date: | 2018-07-05 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Zinc depletion induces ribosome hibernation in mycobacteria.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DZK
 
 | | Cryo-EM Structure of Mycobacterium smegmatis C(minus) 30S ribosomal subunit with MPY | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Sharma, M.R, Li, Y, Korripella, R, Yang, Y, Kaushal, P.S, Lin, Q, Wade, J.T, Gray, A.G, Derbyshire, K.M, Agrawal, R.K, Ojha, A. | | Deposit date: | 2018-07-05 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Zinc depletion induces ribosome hibernation in mycobacteria.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DZI
 
 | | Cryo-EM Structure of Mycobacterium smegmatis 70S C(minus) ribosome 70S-MPY complex | | Descriptor: | 16S rRNA, 23 S rRNA (3119-MER), 30S ribosomal protein S10, ... | | Authors: | Sharma, M.R, Li, Y, Korripella, R, Yang, Y, Kaushal, P.S, Lin, Q, Wade, J.T, Gray, A.G, Derbyshire, K.M, Agrawal, R.K, Ojha, A. | | Deposit date: | 2018-07-05 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Zinc depletion induces ribosome hibernation in mycobacteria.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
