8S6N
 
 | | Structure of MLLE3 in complex with PAMPL1 | | Descriptor: | PAMPL1, RNA-binding protein RRM4 | | Authors: | Devan, S, Shanmugasundaram, S, Muentjes, K, Smits, S.H, Altegoer, F, Feldbruegge, M. | | Deposit date: | 2024-02-28 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.743 Å) | | Cite: | Deciphering the RNA-binding protein network during endosomal mRNA transport
To Be Published
|
|
8S6U
 
 | | Structure of Pab1 in complex with peptide | | Descriptor: | FYVE zinc finger domain protein UPA1, Polyadenylate-binding protein 1 | | Authors: | Devan, S, Shanmugasundaram, S, Muentjes, K, Smits, S.H, Altegoer, F, Feldbruegge, M. | | Deposit date: | 2024-02-28 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Pab1 in complex with peptide
To Be Published
|
|
8S6O
 
 | | Structure of MLLE3 in complex with PAMPL2 | | Descriptor: | PAMPL2, RNA-binding protein RRM4 | | Authors: | Devan, S, Shanmugasundaram, S, Muentjes, K, Smits, S.H, Altegoer, F, Feldbruegge, M. | | Deposit date: | 2024-02-28 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Deciphering the RNA-binding protein network during endosomal mRNA transport
To Be Published
|
|
6QGM
 
 | | VirX1 apo structure | | Descriptor: | VirX1 | | Authors: | Gkotsi, D.S, Ludewig, H, Sharma, S.V, Unsworth, W.P, Taylor, R.J.K, McLachlan, M.M.W, Shanahan, S, Naismith, J.H, Goss, R.J.M. | | Deposit date: | 2019-01-11 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A marine viral halogenase that iodinates diverse substrates.
Nat.Chem., 11, 2019
|
|
8V1S
 
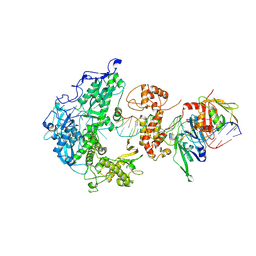 | | Herpes simplex virus 1 polymerase holoenzyme bound to mismatched DNA in editing conformation | | Descriptor: | DNA polymerase, DNA polymerase processivity factor, MAGNESIUM ION, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2023-11-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
8V1T
 
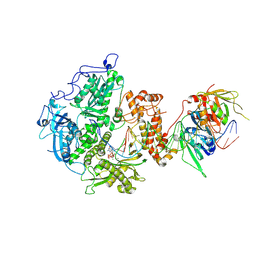 | | Herpes simplex virus 1 polymerase holoenzyme bound to DNA and acyclovir triphosphate in closed conformation | | Descriptor: | ACYCLOVIR TRIPHOSPHATE, DNA polymerase, DNA polymerase processivity factor, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2023-11-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
8V1R
 
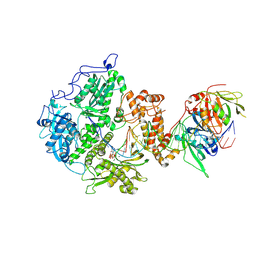 | | Herpes simplex virus 1 polymerase holoenzyme bound to DNA and DTTP in closed conformation | | Descriptor: | DNA polymerase, DNA polymerase processivity factor, MAGNESIUM ION, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2023-11-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
8V1Q
 
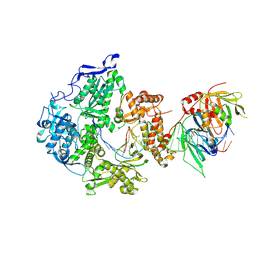 | | Herpes simplex virus 1 polymerase holoenzyme bound to DNA in both open/closed conformations | | Descriptor: | DNA polymerase, DNA polymerase processivity factor, MAGNESIUM ION, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2023-11-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
6FAL
 
 | |
6F7F
 
 | |
6F7G
 
 | |
7YK5
 
 | | Rubisco from Phaeodactylum tricornutum bound to PYCO1(452-592) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, Multifunctional fusion protein, PYCO1 LSU binding motif, ... | | Authors: | Oh, Z.G, Ang, W.S.L, Bhushan, S, Mueller-Cajar, O. | | Deposit date: | 2022-07-21 | | Release date: | 2023-06-21 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | A linker protein from a red-type pyrenoid phase separates with Rubisco via oligomerizing sticker motifs.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6F9K
 
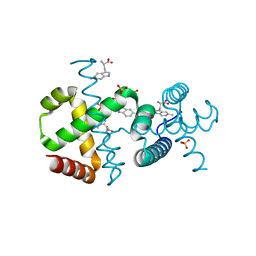 | |
3JYI
 
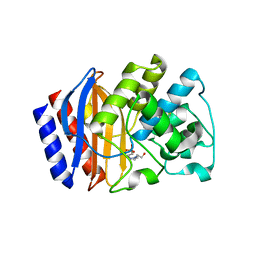 | | Structural and biochemical evidence that a TEM-1 {beta}-lactamase Asn170Gly active site mutant acts via substrate-assisted catalysis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase TEM, PHOSPHATE ION | | Authors: | Brown, N.G, Palzkill, T.G, Prasad, B.V.V, Shanker, S. | | Deposit date: | 2009-09-21 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structural and biochemical evidence that a TEM-1 beta-lactamase N170G active site mutant acts via substrate-assisted catalysis
J.Biol.Chem., 284, 2009
|
|
1T61
 
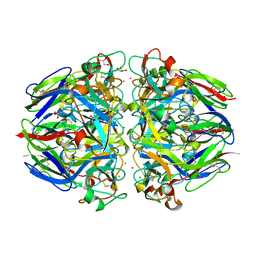 | | crystal structure of collagen IV NC1 domain from placenta basement membrane | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Vanacore, R.M, Shanmugasundararaj, S, Friedman, D.B, Bondar, O, Hudson, B.G, Sundaramoorthy, M. | | Deposit date: | 2004-05-05 | | Release date: | 2004-09-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The alpha1.alpha2 network of collagen IV. Reinforced stabilization of the noncollagenous domain-1 by noncovalent forces and the absence of Met-Lys cross-links
J.Biol.Chem., 279, 2004
|
|
3IZD
 
 | | Model of the large subunit RNA expansion segment ES27L-out based on a 6.1 A cryo-EM map of Saccharomyces cerevisiae translating 80S ribosome. 3IZD is a small part (an expansion segment) which is in an alternative conformation to what is in already 3IZF. | | Descriptor: | rRNA expansion segment ES27L in an "out" conformation | | Authors: | Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | Deposit date: | 2010-10-13 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1T60
 
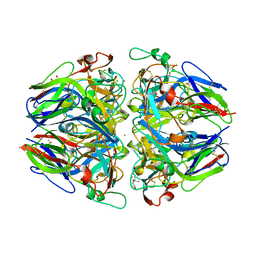 | | Crystal structure of Type IV collagen NC1 domain from bovine lens capsule | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Vanacore, R.M, Shanmugasundararaj, S, Friedman, D.B, Bondar, O, Hudson, B.G, Sundaramoorthy, M. | | Deposit date: | 2004-05-05 | | Release date: | 2004-09-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The alpha1.alpha2 network of collagen IV. Reinforced stabilization of the noncollagenous domain-1 by noncovalent forces and the absence of Met-Lys cross-links
J.Biol.Chem., 279, 2004
|
|
8EXX
 
 | | Herpes simplex virus 1 polymerase holoenzyme bound to DNA and foscarnet (pre-translocation state) | | Descriptor: | DNA polymerase, DNA polymerase processivity factor, MAGNESIUM ION, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2022-10-26 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
6JK2
 
 | | Crystal structure of a mini fungal lectin, PhoSL | | Descriptor: | Lectin, SULFATE ION | | Authors: | Lou, Y.C, Chou, C.C, Yeh, H.H, Chien, C.Y, Sushant, S, Chen, C, Hsu, C.H. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Structural insights into the role of N-terminal integrity in PhoSL for core-fucosylated N-glycan recognition.
Int.J.Biol.Macromol., 255, 2023
|
|
6SLD
 
 | | Structure of the Phosphatidylcholine Binding Mutant of Yeast Sec14 Homolog Sfh1 (S175I,T177I) in Complex with Phosphatidylinositol | | Descriptor: | (1R)-2-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CRAL-TRIO domain-containing protein YKL091C | | Authors: | Berger, J, Fitz, M, Johnen, P, Shanmugaratnam, S, Hocker, B, Stiel, A.C, Schaaf, G. | | Deposit date: | 2019-08-19 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the Phosphatidylcholine Binding Mutant of Yeast Sec14 Homolog Sfh1 (S175I,T177I) in Complex with Phosphatidylinositol
To Be Published
|
|
7RCU
 
 | | Synthetic Max homodimer mimic in complex with DNA | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, 2-(2,5-dioxopyrrolidin-1-yl)acetamide, ACETAMIDE, ... | | Authors: | Speltz, T, Qiao, Z, Shangguan, S, Fanning, S, Greene, J, Moellering, R. | | Deposit date: | 2021-07-08 | | Release date: | 2022-09-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Targeting MYC with modular synthetic transcriptional repressors derived from bHLH DNA-binding domains.
Nat.Biotechnol., 41, 2023
|
|
6JK3
 
 | | Crystal structure of a mini fungal lectin, PhoSL in complex with core-fucosylated chitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Lectin | | Authors: | Lou, Y.C, Chou, C.C, Yeh, H.H, Chien, C.Y, Sushant, S, Chen, C, Hsu, C.H. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural insights into the role of N-terminal integrity in PhoSL for core-fucosylated N-glycan recognition.
Int.J.Biol.Macromol., 255, 2023
|
|
5ZEY
 
 | | M. smegmatis Trans-translation state 70S ribosome | | Descriptor: | A-tRNAfMet, SsrA-binding protein, tmRNA | | Authors: | Mishra, S, Ahmed, T, Tyagi, A, Shi, J, Bhushan, S. | | Deposit date: | 2018-02-28 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (12.5 Å) | | Cite: | Structures of Mycobacterium smegmatis 70S ribosomes in complex with HPF, tmRNA, and P-tRNA.
Sci Rep, 8, 2018
|
|
5ZEB
 
 | | M. Smegmatis P/P state 70S ribosome structure | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Mishra, S, Ahmed, T, Tyagi, A, Shi, J, Bhushan, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-09-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Mycobacterium smegmatis 70S ribosomes in complex with HPF, tmRNA, and P-tRNA.
Sci Rep, 8, 2018
|
|
5A9Z
 
 | | Complex of Thermous thermophilus ribosome bound to BipA-GDPCP | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Kumar, V, Chen, Y, Ahmed, T, Tan, J, Ero, R, Bhushan, S, Gao, Y.-G. | | Deposit date: | 2015-07-23 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
