1GBN
 
 | | HUMAN ORNITHINE AMINOTRANSFERASE COMPLEXED WITH THE NEUROTOXIN GABACULINE | | Descriptor: | 3-AMINOBENZOIC ACID, GABACULINE, ORNITHINE AMINOTRANSFERASE, ... | | Authors: | Shah, S.A, Shen, B.W, Brunger, A.T. | | Deposit date: | 1997-05-29 | | Release date: | 1998-06-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human ornithine aminotransferase complexed with L-canaline and gabaculine: structural basis for substrate recognition.
Structure, 5, 1997
|
|
4WA8
 
 | |
406D
 
 | | 5'-R(*CP*AP*CP*CP*GP*GP*AP*UP*GP*GP*UP*(BRO) UP*CP*GP*GP*UP*G)-3' | | Descriptor: | RNA (5'-R(*CP*AP*CP*CP*GP*GP*AP*UP*GP*GP*UP*(5BU)P*CP*GP*GP*UP*G)-3') | | Authors: | Shah, S.A, Brunger, A.T. | | Deposit date: | 1998-06-22 | | Release date: | 1999-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8 A crystal structure of a statically disordered 17 base-pair RNA duplex: principles of RNA crystal packing and its effect on nucleic acid structure.
J.Mol.Biol., 285, 1999
|
|
2CAN
 
 | |
9FSS
 
 | | RNA Polymerase III Class III Open Mini Pre-Initiation complex 2 (OC2-mini) | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Shah, S.Z, Ramsay, E.P, Cecatiello, V, Perry, T.N, Vannini, A. | | Deposit date: | 2024-06-21 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Structural insights into distinct mechanisms of RNA polymerase II and III recruitment to snRNA promoters.
Nat Commun, 16, 2025
|
|
9FSO
 
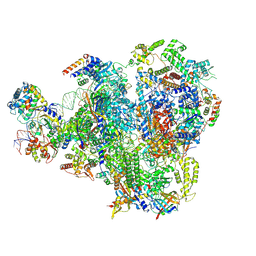 | | Human RNA Polymerase III Class III Open Pre-initiation Complex 1 (OC1) | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Shah, S.Z, Ramsay, E.P, Cecatiello, V, Perry, T.N, Vannini, A. | | Deposit date: | 2024-06-21 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural insights into distinct mechanisms of RNA polymerase II and III recruitment to snRNA promoters.
Nat Commun, 16, 2025
|
|
9FSP
 
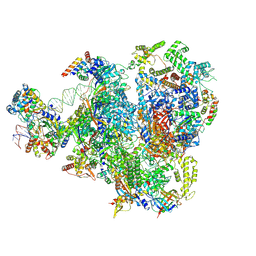 | | RNA Polymerase III Class III Open Pre-initation Complex 2 (OC2) | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Shah, S.Z, Ramsay, E.P, Cecatiello, V, Perry, T.N, Vannini, A. | | Deposit date: | 2024-06-21 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural insights into distinct mechanisms of RNA polymerase II and III recruitment to snRNA promoters.
Nat Commun, 16, 2025
|
|
9FSR
 
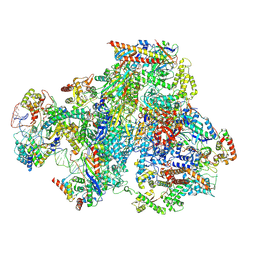 | | RNA Polymerase III Class III Open Mini Pre-initiation Complex 1 (OC1-mini) | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Shah, S.Z, Ramsay, E.P, Cecatiello, V, Perry, T.N, Vannini, A. | | Deposit date: | 2024-06-21 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural insights into distinct mechanisms of RNA polymerase II and III recruitment to snRNA promoters.
Nat Commun, 16, 2025
|
|
9FSQ
 
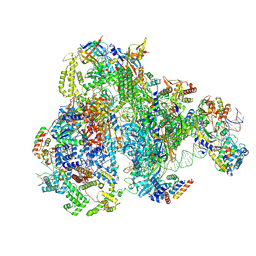 | | RNA Polymerase III Class III Melting Pre-Initiation Complex (MC) | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Shah, S.Z, Ramsay, E.P, Cecatiello, V, Perry, T.N, Vannini, A. | | Deposit date: | 2024-06-21 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural insights into distinct mechanisms of RNA polymerase II and III recruitment to snRNA promoters.
Nat Commun, 16, 2025
|
|
2LME
 
 | | Solid-state NMR structure of the membrane anchor domain of the trimeric autotransporter YadA | | Descriptor: | Adhesin yadA | | Authors: | Shahid, S.A, Bardiaux, B, Franks, W.T, Habeck, M, Linke, D, van Rossum, B. | | Deposit date: | 2011-11-30 | | Release date: | 2012-11-07 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Membrane-protein structure determination by solid-state NMR spectroscopy of microcrystals.
Nat.Methods, 9, 2012
|
|
8TFE
 
 | |
6X9X
 
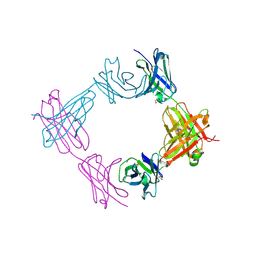 | |
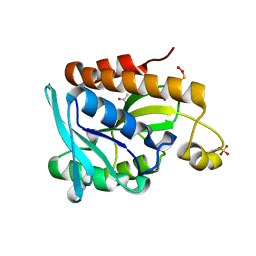
5GVZ
 
 | |
5IVP
 
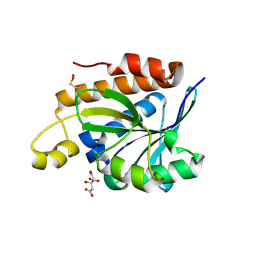 | |
5ZK0
 
 | | Crystal structure of Peptidyl-tRNA hydrolase mutant -M71A from Vibrio cholerae | | Descriptor: | Peptidyl-tRNA hydrolase, SODIUM ION | | Authors: | Shahid, S, Kabra, A, Pal, R.K, Arora, A. | | Deposit date: | 2018-03-22 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Role of methionine 71 in substrate recognition and structural integrity of bacterial peptidyl-tRNA hydrolase.
Biochim. Biophys. Acta, 1866, 2018
|
|
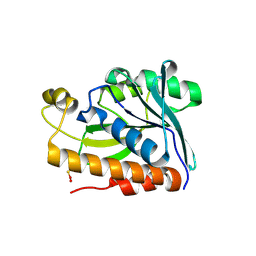
5EKT
 
 | |
8TGV
 
 | | CryoEM structure of Fab HC84.26-HCV E2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HC84.26 Heavy chain, ... | | Authors: | Shahid, S, Liqun, J, LIu, Y, Hasan, S.S, Mariuzza, R.A. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | CryoEM structure of Fab HC84.26-HCV E2 complex
To Be Published
|
|
8TGZ
 
 | | CryoEM structure of neutralizing antibody HC84.26 in complex with Hepatitis C virus envelope glycoprotein E2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HC84.26 Heavy chain, ... | | Authors: | Shahid, S, Liqun, J, Liu, Y, Hasan, S.S, Mariuzza, R.A. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | CryoEM structure of neutralizing antibody HC84.26 in complex with Hepatitis C virus envelope glycoprotein E2
To Be Published
|
|
8THZ
 
 | | CryoEM structure of neutralizing antibodies CBH-7 and HC84.26 in complex with Hepatitis C virus envelope glycoprotein E2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CBH-7 Heavy chain, ... | | Authors: | Shahid, S, Jiang, L, Liu, Y, Hasan, S.S, Mariuzza, R.A. | | Deposit date: | 2023-07-18 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | CryoEM structure of neutralizing antibodies CBH-7 and HC84.26 in complex with Hepatitis C virus envelope glycoprotein E2
To Be Published
|
|
8TXQ
 
 | | CryoEM structure of non-neutralizing antibody CBH-4B in complex with Hepatitis C virus envelope glycoprotein E2 | | Descriptor: | CBH-4B Heavy chain, CBH-4B Light chain, envelope glycoprotein E2 | | Authors: | Shahid, S, Liqun, J, Liu, Y, Hasan, S.S, Mariuzza, R.A. | | Deposit date: | 2023-08-24 | | Release date: | 2024-08-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | CryoEM structure of non-neutralizing antibody CBH-4B in complex with Hepatitis C virus envelope glycoprotein E2
To Be Published
|
|
8TZY
 
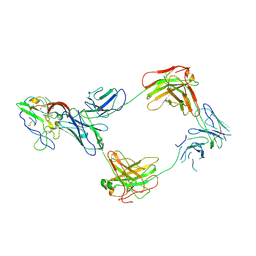 | | CryoEM structure of non-neutralizing bivalent antibody CBH-4B in complex with Hepatitis C virus envelope glycoprotein E2 | | Descriptor: | CBH4B Heavy chain, CBH4B Light chain, envelope glycoprotein E2 | | Authors: | Shahid, S, Liqun, J, Liu, Y, Hasan, S.S, Mariuzza, R.A. | | Deposit date: | 2023-08-28 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | CryoEM structure of non-neutralizing bivalent antibody CBH-4B in complex with Hepatitis C virus envelope glycoprotein E2
To Be Published
|
|
8U9Y
 
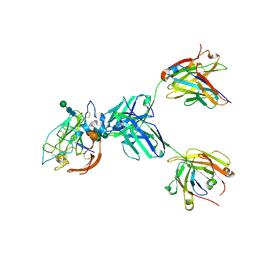 | | CryoEM structure of neutralizing antibody HC84.26 in complex with Hepatitis C virus envelope glycoprotein E2_New interface | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | Authors: | Shahid, S, Liqun, J, Liu, Y, Hasan, S.S, Mariuzza, R.A. | | Deposit date: | 2023-09-20 | | Release date: | 2024-09-25 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | CryoEM structure of neutralizing antibody HC84.26 in complex with Hepatitis C virus envelope glycoprotein E2_New interface
To Be Published
|
|
5IKE
 
 | |
5IMB
 
 | |
4Z86
 
 | |
