6O0P
 
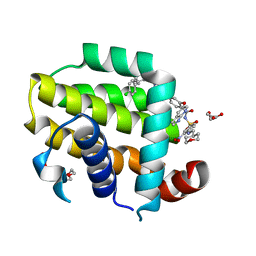 | | crystal structure of BCL-2 G101A mutation with venetoclax | | Descriptor: | 4-{4-[(4'-chloro-5,5-dimethyl[3,4,5,6-tetrahydro[1,1'-biphenyl]]-2-yl)methyl]piperazin-1-yl}-N-[(3-nitro-4-{[(oxan-4-yl )methyl]amino}phenyl)sulfonyl]-2-[(1H-pyrrolo[2,3-b]pyridin-5-yl)oxy]benzamide, Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2, DI(HYDROXYETHYL)ETHER | | Authors: | Birkinshaw, R.W, Luo, C.S, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-02-17 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Nat Commun, 10, 2019
|
|
6O0M
 
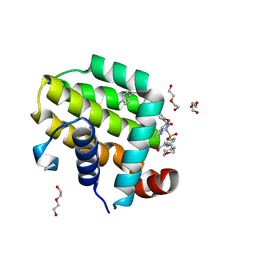 | | crystal structure of BCL-2 F104L mutation with venetoclax | | Descriptor: | 4-{4-[(4'-chloro-5,5-dimethyl[3,4,5,6-tetrahydro[1,1'-biphenyl]]-2-yl)methyl]piperazin-1-yl}-N-[(3-nitro-4-{[(oxan-4-yl )methyl]amino}phenyl)sulfonyl]-2-[(1H-pyrrolo[2,3-b]pyridin-5-yl)oxy]benzamide, Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2, DI(HYDROXYETHYL)ETHER | | Authors: | Birkinshaw, R.W, Luo, C.S, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-02-16 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Nat Commun, 10, 2019
|
|
6O0K
 
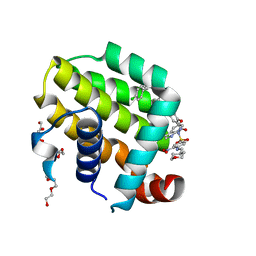 | | crystal structure of BCL-2 with venetoclax | | Descriptor: | 4-{4-[(4'-chloro-5,5-dimethyl[3,4,5,6-tetrahydro[1,1'-biphenyl]]-2-yl)methyl]piperazin-1-yl}-N-[(3-nitro-4-{[(oxan-4-yl )methyl]amino}phenyl)sulfonyl]-2-[(1H-pyrrolo[2,3-b]pyridin-5-yl)oxy]benzamide, Apoptosis regulator Bcl-2, NONAETHYLENE GLYCOL | | Authors: | Birkinshaw, R.W, Luo, C.S, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-02-16 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Nat Commun, 10, 2019
|
|
2X8L
 
 | |
7K02
 
 | |
7LV3
 
 | | Crystal structure of human protein kinase G (PKG) R-C complex in inhibited state | | Descriptor: | 1,2-ETHANEDIOL, Isoform Beta of cGMP-dependent protein kinase 1, MANGANESE (II) ION, ... | | Authors: | Sharma, R, Lying, Q, Casteel, D, Kim, C. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | An auto-inhibited state of protein kinase G and implications for selective activation.
Elife, 11, 2022
|
|
1QF3
 
 | | PEANUT LECTIN COMPLEXED WITH METHYL-BETA-GALACTOSE | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PROTEIN (PEANUT LECTIN), ... | | Authors: | Ravishankar, R, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 1999-04-06 | | Release date: | 1999-07-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the complexes of peanut lectin with methyl-beta-galactose and N-acetyllactosamine and a comparative study of carbohydrate binding in Gal/GalNAc-specific legume lectins.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BZW
 
 | | PEANUT LECTIN COMPLEXED WITH C-LACTOSE | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PROTEIN (PEANUT LECTIN), ... | | Authors: | Ravishankar, R, Surolia, A, Vijayan, M, Lim, S, Kishi, Y. | | Deposit date: | 1998-11-05 | | Release date: | 1998-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Preferred Conformation of C-Lactose at the Free and Peanut Lectin Bound States
J.Am.Chem.Soc., 120, 1998
|
|
1YYE
 
 | | Crystal structure of estrogen receptor beta complexed with way-202196 | | Descriptor: | 3-(3-FLUORO-4-HYDROXYPHENYL)-7-HYDROXY-1-NAPHTHONITRILE, Estrogen receptor beta, STEROID RECEPTOR COACTIVATOR-1 | | Authors: | Mewshaw, R.E, Edsall Jr, R.J, Yang, C, Manas, E.S, Xu, Z.B, Henderson, R.A, Keith Jr, J.C, Harris, H.A. | | Deposit date: | 2005-02-24 | | Release date: | 2006-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | ERbeta ligands. 3. Exploiting two binding orientations of the 2-phenylnaphthalene scaffold to achieve ERbeta selectivity
J.Med.Chem., 48, 2005
|
|
1CR7
 
 | | PEANUT LECTIN-LACTOSE COMPLEX MONOCLINIC FORM | | Descriptor: | CALCIUM ION, LECTIN, MANGANESE (II) ION, ... | | Authors: | Ravishankar, R, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 1999-08-14 | | Release date: | 2001-04-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the peanut lectin-lactose complex at acidic pH: retention of unusual quaternary structure, empty and carbohydrate bound combining sites, molecular mimicry and crystal packing directed by interactions at the combining site.
Proteins, 43, 2001
|
|
8QHC
 
 | | Cryo-EM structure of SidH from Legionella pneumophila in complex with LubX | | Descriptor: | E3 ubiquitin--protein ligase, Elongation factor Tu, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Sharma, R, Adams, M, Bhogaraju, S. | | Deposit date: | 2023-09-07 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the toxicity of Legionella pneumophila effector SidH.
Nat Commun, 14, 2023
|
|
8QFS
 
 | | Cryo-EM structure of SidH from Legionella pneumophila | | Descriptor: | Elongation factor Tu, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sharma, R, Weis, F, Bhogaraju, S. | | Deposit date: | 2023-09-04 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for the toxicity of Legionella pneumophila effector SidH.
Nat Commun, 14, 2023
|
|
1YY4
 
 | | Crystal structure of estrogen receptor beta complexed with 1-chloro-6-(4-hydroxy-phenyl)-naphthalen-2-ol | | Descriptor: | 1-CHLORO-6-(4-HYDROXYPHENYL)-2-NAPHTHOL, Estrogen receptor beta, STEROID RECEPTOR COACTIVATOR-1 | | Authors: | Mewshaw, R.E, Edsall Jr, R.J, Yang, C, Manas, E.S, Xu, Z.B, Henderson, R.A, Keith Jr, J.C, Harris, H.A. | | Deposit date: | 2005-02-23 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | ERbeta ligands. 3. Exploiting two binding orientations of the 2-phenylnaphthalene scaffold to achieve ERbeta selectivity
J.Med.Chem., 48, 2005
|
|
1CIW
 
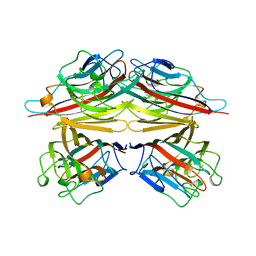 | | PEANUT LECTIN COMPLEXED WITH N-ACETYLLACTOSAMINE | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PROTEIN (PEANUT LECTIN), ... | | Authors: | Ravishankar, R, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 1999-04-06 | | Release date: | 1999-07-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of the complexes of peanut lectin with methyl-beta-galactose and N-acetyllactosamine and a comparative study of carbohydrate binding in Gal/GalNAc-specific legume lectins.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1EUI
 
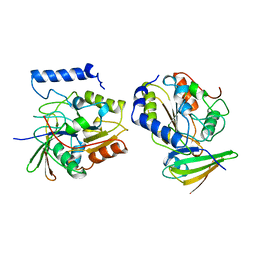 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE COMPLEX WITH URACIL-DNA GLYCOSYLASE INHIBITOR PROTEIN | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR PROTEIN | | Authors: | Ravishankar, R, Sagar, M.B, Roy, S, Purnapatre, K, Handa, P, Varshney, U, Vijayan, M. | | Deposit date: | 1998-06-18 | | Release date: | 1999-06-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray analysis of a complex of Escherichia coli uracil DNA glycosylase (EcUDG) with a proteinaceous inhibitor. The structure elucidation of a prokaryotic UDG.
Nucleic Acids Res., 26, 1998
|
|
1CKR
 
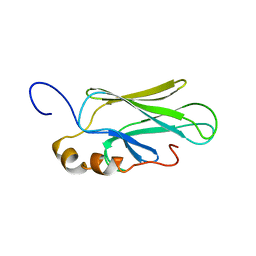 | | HIGH RESOLUTION SOLUTION STRUCTURE OF THE HEAT SHOCK COGNATE-70 KD SUBSTRATE BINDING DOMAIN OBTAINED BY MULTIDIMENSIONAL NMR TECHNIQUES | | Descriptor: | HEAT SHOCK SUBSTRATE BINDING DOMAIN OF HSC-70 | | Authors: | Morshauser, R.C, Hu, W, Wang, H, Pang, Y, Flynn, G.C, Zuiderweg, E.R.P. | | Deposit date: | 1999-04-22 | | Release date: | 1999-04-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of the 18 kDa substrate-binding domain of the mammalian chaperone protein Hsc70.
J.Mol.Biol., 289, 1999
|
|
1CQ9
 
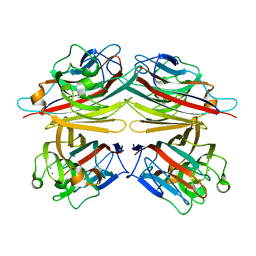 | | PEANUT LECTIN-TRICLINIC FORM | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PROTEIN (PEANUT LECTIN) | | Authors: | Ravishankar, R, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 1999-08-06 | | Release date: | 2002-05-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of the peanut lectin-lactose complex at acidic pH: retention of unusual
quaternary structure, empty and carbohydrate bound combining sites, molecular mimicry
and crystal packing directed by interactions at the combining site.
Proteins, 43, 2001
|
|
6UX8
 
 | |
7L99
 
 | | Crystal structure of BRDT bromodomain 2 in complex with CDD-1302 | | Descriptor: | Bromodomain testis-specific protein, N-[3-(acetylamino)-4-methylphenyl]-3-(4-amino-2-methylphenyl)-1-methyl-1H-indazole-5-carboxamide, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500) | | Authors: | Sharma, R, Yu, Z, Ku, A.F, Anglin, J.L, Ucisik, M.N, Faver, J.C, Sankaran, B, Kim, C, Matzuk, M.M. | | Deposit date: | 2021-01-03 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and characterization of bromodomain 2-specific inhibitors of BRDT.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7L9A
 
 | | Crystal structure of BRDT bromodomain 2 in complex with CDD-1102 | | Descriptor: | BETA-MERCAPTOETHANOL, Bromodomain testis-specific protein, N~1~-(5-{[3-(4-amino-2-methylphenyl)-1-methyl-1H-indazole-5-carbonyl]amino}-2-methylphenyl)-N~4~-methylbenzene-1,4-dicarboxamide | | Authors: | Sharma, R, Kaur, G, Yu, Z, Ku, A.F, Anglin, J.L, Ucisik, M.N, Faver, J.C, Sankaran, B, Kim, C, Matzuk, M.M. | | Deposit date: | 2021-01-03 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery and characterization of bromodomain 2-specific inhibitors of BRDT.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4X6C
 
 | | CD1a ternary complex with lysophosphatidylcholine and BK6 TCR | | Descriptor: | (4R,7R,18Z)-4,7-dihydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphaheptacos-18-en-1-aminium 4-oxide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2014-12-08 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | alpha beta T cell antigen receptor recognition of CD1a presenting self lipid ligands.
Nat.Immunol., 16, 2015
|
|
4X6D
 
 | | CD1a ternary complex with endogenous lipids and BK6 TCR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, OLEIC ACID, ... | | Authors: | Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2014-12-08 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | alpha beta T cell antigen receptor recognition of CD1a presenting self lipid ligands.
Nat.Immunol., 16, 2015
|
|
4X6F
 
 | | CD1a binary complex with sphingomyelin | | Descriptor: | (4S,7S,23Z)-4-hydroxy-7-[(1S,2Z)-1-hydroxyhexadec-2-en-1-yl]-N,N,N-trimethyl-9-oxo-3,5-dioxa-8-aza-4-phosphadotriacont- 23-en-1-aminium 4-oxide, Beta-2-microglobulin, T-cell surface glycoprotein CD1a | | Authors: | Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2014-12-08 | | Release date: | 2015-02-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | alpha beta T cell antigen receptor recognition of CD1a presenting self lipid ligands.
Nat.Immunol., 16, 2015
|
|
5H23
 
 | | Crystal structure of Chikungunya virus capsid protein | | Descriptor: | 1,2-ETHANEDIOL, Capsid Protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sharma, R, Kesari, P, Tomar, S, Kumar, P. | | Deposit date: | 2016-10-14 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-function insights into chikungunya virus capsid protein: Small molecules targeting capsid hydrophobic pocket.
Virology, 515, 2018
|
|
4X6E
 
 | | CD1a binary complex with lysophosphatidylcholine | | Descriptor: | (4R,7R,18Z)-4,7-dihydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphaheptacos-18-en-1-aminium 4-oxide, Beta-2-microglobulin, D-MALATE, ... | | Authors: | Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2014-12-08 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | alpha beta T cell antigen receptor recognition of CD1a presenting self lipid ligands.
Nat.Immunol., 16, 2015
|
|
