6AVI
 
 | |
6DGP
 
 | |
7JQG
 
 | |
6VW1
 
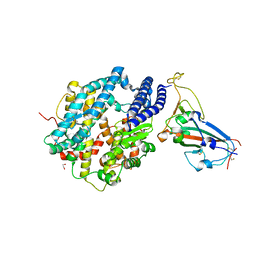 | | Structure of SARS-CoV-2 chimeric receptor-binding domain complexed with its receptor human ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shang, J, Ye, G, Shi, K, Wan, Y.S, Aihara, H, Li, F. | | Deposit date: | 2020-02-18 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural basis of receptor recognition by SARS-CoV-2.
Nature, 581, 2020
|
|
6B7N
 
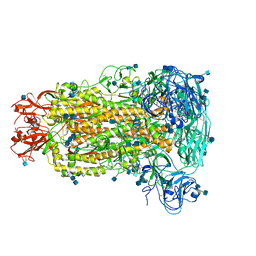 | | Cryo-electron microscopy structure of porcine delta coronavirus spike protein in the pre-fusion state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shang, J, Zheng, Y, Yang, Y, Liu, C, Geng, Q, Tai, W, Du, L, Zhou, Y, Zhang, W, Li, F. | | Deposit date: | 2017-10-04 | | Release date: | 2017-10-25 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-Electron Microscopy Structure of Porcine Deltacoronavirus Spike Protein in the Prefusion State
J. Virol., 92, 2018
|
|
8HG6
 
 | | Cryo-EM structure of the prasinophyte-specific light-harvesting complex (Lhcp)from Ostreococcus tauri | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (1~{S})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaenyl]cyclohex-3-en-1-ol, (3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-1-[(1~{S},4~{S})-2,2-dimethyl-6-methylidene-1,4-bis(oxidanyl)cyclohexyl]-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaen-2-one, ... | | Authors: | Shan, J, Sheng, X, Ishii, A, Watanabe, A, Song, C, Murata, K, Minagawa, J, Liu, Z. | | Deposit date: | 2022-11-13 | | Release date: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | The photosystem I supercomplex from a primordial green alga Ostreococcus tauri harbors three light-harvesting complex trimers.
Elife, 12, 2023
|
|
8HG5
 
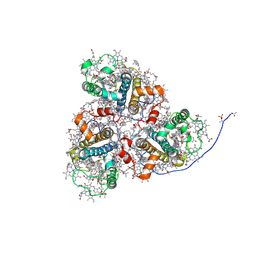 | | Cryo-EM structure of the prasinophyte-specific light-harvesting complex (Lhcp)from Ostreococcus tauri | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (1~{S})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaenyl]cyclohex-3-en-1-ol, (3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-1-[(1~{S},4~{S})-2,2-dimethyl-6-methylidene-1,4-bis(oxidanyl)cyclohexyl]-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaen-2-one, ... | | Authors: | Shan, J, Sheng, X, Ishii, A, Watanabe, A, Song, C, Murata, K, Minagawa, J, Liu, Z. | | Deposit date: | 2022-11-13 | | Release date: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The photosystem I supercomplex from a primordial green alga Ostreococcus tauri harbors three light-harvesting complex trimers.
Elife, 12, 2023
|
|
8HG3
 
 | | Cryo-EM structure of the Lhcp complex from Ostreococcus tauri | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (1~{S})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaenyl]cyclohex-3-en-1-ol, (3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-1-[(1~{S},4~{S})-2,2-dimethyl-6-methylidene-1,4-bis(oxidanyl)cyclohexyl]-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaen-2-one, ... | | Authors: | Shan, J, Sheng, X, Ishii, A, Watanabe, A, Song, C, Murata, K, Minagawa, J, Liu, Z. | | Deposit date: | 2022-11-13 | | Release date: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | The photosystem I supercomplex from a primordial green alga Ostreococcus tauri harbors three light-harvesting complex trimers.
Elife, 12, 2023
|
|
6CV0
 
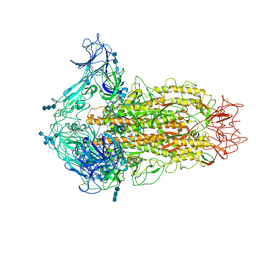 | | Cryo-electron microscopy structure of infectious bronchitis coronavirus spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shang, J, Zheng, Y, Yang, Y, Liu, C, Geng, Q, Luo, C, Zhang, W, Li, F. | | Deposit date: | 2018-03-27 | | Release date: | 2018-04-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Cryo-EM structure of infectious bronchitis coronavirus spike protein reveals structural and functional evolution of coronavirus spike proteins.
PLoS Pathog., 14, 2018
|
|
5CR6
 
 | | Structure of pneumolysin at 1.98 A resolution | | Descriptor: | Pneumolysin | | Authors: | Marshall, J.E, Faraj, B.H.A, Gingras, A.R, Lonnen, R, Sheikh, M.A, El-Mezgueldi, M, Moody, P.C.E, Andrew, P.W, Wallis, R. | | Deposit date: | 2015-07-22 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Crystal Structure of Pneumolysin at 2.0 angstrom Resolution Reveals the Molecular Packing of the Pre-pore Complex.
Sci Rep, 5, 2015
|
|
5CR8
 
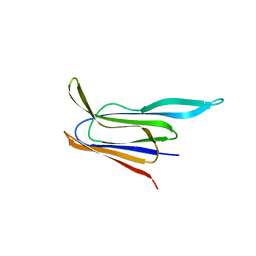 | | Structure of the membrane-binding domain of pneumolysin | | Descriptor: | Pneumolysin | | Authors: | Marshall, J.E, Faraj, B.H.A, Gingras, A.R, Lonnen, R, Sheikh, M.A, El-Mezgueldi, M, Moody, P.C.E, Andrew, P.W, Wallis, R. | | Deposit date: | 2015-07-22 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Crystal Structure of Pneumolysin at 2.0 angstrom Resolution Reveals the Molecular Packing of the Pre-pore Complex.
Sci Rep, 5, 2015
|
|
7L5P
 
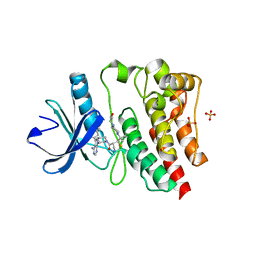 | | Crystal structure of the covalently bonded complex of rilzabrutinib with BTK | | Descriptor: | (2E)-2-{(3R)-3-[4-amino-3-(2-fluoro-4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidine-1-carbonyl}-4-methyl-4-[4-(oxetan-3-yl)piperazin-1-yl]pent-2-enenitrile, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Bradshaw, J.M, Brameld, K.A, Mrosek, M, Lammens, A, Blaesse, M. | | Deposit date: | 2020-12-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The Discovery Rilzabrutinib (PRN1008): A Reversible Covalent BTK Inhibitor for Immune Mediated Diseases
To Be Published
|
|
2CDO
 
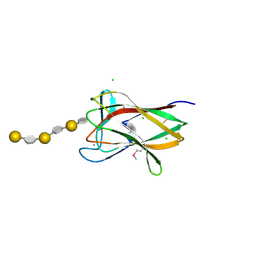 | | structure of agarase carbohydrate binding module in complex with neoagarohexaose | | Descriptor: | 1,2-ETHANEDIOL, 3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose, BETA-AGARASE 1, ... | | Authors: | Henshaw, J, Horne, A, Van Bueren, A.L, Money, V.A, Bolam, D.N, Czjzek, M, Weiner, R.M, Hutcheson, S.W, Davies, G.J, Boraston, A.B, Gilbert, H.J. | | Deposit date: | 2006-01-25 | | Release date: | 2006-02-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Family 6 Carbohydrate Binding Modules in Beta-Agarases Display Exquisite Selectivity for the Non- Reducing Termini of Agarose Chains.
J.Biol.Chem., 281, 2006
|
|
2CDP
 
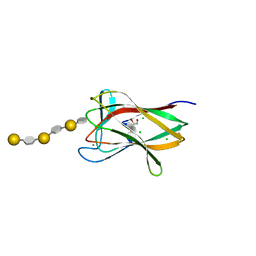 | | Structure of a CBM6 in complex with neoagarohexaose | | Descriptor: | 1,2-ETHANEDIOL, 3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose, BETA-AGARASE 1, ... | | Authors: | Henshaw, J, Horne, A, Van Bueren, A.L, Money, V.A, Bolam, D.N, Czjzek, M, Weiner, R.M, Hutcheson, S.W, Davies, G.J, Boraston, A.B, Gilbert, H.J. | | Deposit date: | 2006-01-26 | | Release date: | 2006-02-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Family 6 Carbohydrate Binding Modules in Beta-Agarases Display Exquisite Selectivity for the Non- Reducing Termini of Agarose Chains.
J.Biol.Chem., 281, 2006
|
|
3FPU
 
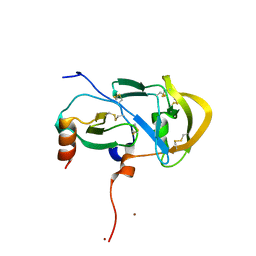 | |
3FPT
 
 | | The Crystal Structure of the Complex between Evasin-1 and CCL3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Evasin-1 | | Authors: | Shaw, J.P, Dias, J.M. | | Deposit date: | 2009-01-06 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of chemokine sequestration by a tick chemokine binding protein: the crystal structure of the complex between Evasin-1 and CCL3
Plos One, 4, 2009
|
|
5UGM
 
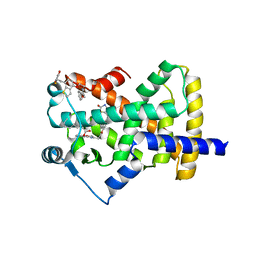 | | Crystal Structure of Human PPARgamma Ligand Binding Domain in Complex with Edaglitazone | | Descriptor: | (5R)-5-({4-[2-(5-methyl-2-phenyl-1,3-oxazol-4-yl)ethoxy]-1-benzothiophen-7-yl}methyl)-1,3-thiazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma, nonanoic acid | | Authors: | Shang, J, Kojetin, D.J. | | Deposit date: | 2017-01-09 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cooperative cobinding of synthetic and natural ligands to the nuclear receptor PPAR gamma.
Elife, 7, 2018
|
|
1SFT
 
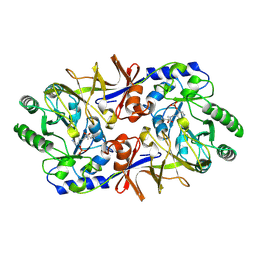 | | ALANINE RACEMASE | | Descriptor: | ACETATE ION, ALANINE RACEMASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Shaw, J.P, Petsko, G.A, Ringe, D. | | Deposit date: | 1996-09-20 | | Release date: | 1997-02-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determination of the structure of alanine racemase from Bacillus stearothermophilus at 1.9-A resolution.
Biochemistry, 36, 1997
|
|
1U4R
 
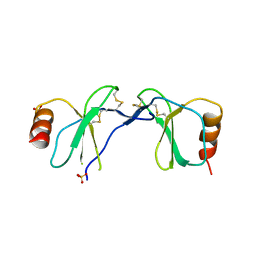 | | Crystal Structure of human RANTES mutant 44-AANA-47 | | Descriptor: | SULFATE ION, Small inducible cytokine A5 | | Authors: | Shaw, J.P, Johnson, Z, Borlat, F, Zwahlen, C, Kungl, A, Roulin, K, Harrenga, A, Wells, T.N.C, Proudfoot, A.E.I. | | Deposit date: | 2004-07-26 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The X-ray structure of RANTES: heparin-derived disaccharides allows the rational design of chemokine inhibitors.
Structure, 12, 2004
|
|
7L5O
 
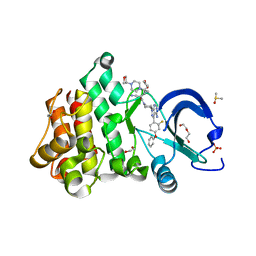 | | Crystal structure of the noncovalently bonded complex of rilzabrutinib with BTK | | Descriptor: | (2E)-2-{(3R)-3-[4-amino-3-(2-fluoro-4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidine-1-carbonyl}-4-methyl-4-[4-(oxetan-3-yl)piperazin-1-yl]pent-2-enenitrile, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Bradshaw, J.M, Brameld, K.A, Mrosek, M, Lammens, A, Blaesse, M. | | Deposit date: | 2020-12-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | The Discovery Rilzabrutinib (PRN1008): A Reversible Covalent BTK Inhibitor for Immune Mediated Diseases
To Be Published
|
|
7YCA
 
 | | Cryo-EM structure of the PSI-LHCI-Lhcp supercomplex from Ostreococcus tauri | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (1~{S})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaenyl]cyclohex-3-en-1-ol, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Shan, J, Sheng, X, Ishii, A, Watanabe, A, Song, C, Murata, K, Minagawa, J, Liu, Z. | | Deposit date: | 2022-07-01 | | Release date: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | The photosystem I supercomplex from a primordial green alga Ostreococcus tauri harbors three light-harvesting complex trimers.
Elife, 12, 2023
|
|
1U4L
 
 | | human RANTES complexed to heparin-derived disaccharide I-S | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ACETIC ACID, Small inducible cytokine A5 | | Authors: | Shaw, J.P, Johnson, Z, Borlat, F, Zwahlen, C, Kungl, A, Roulin, K, Harrenga, A, Wells, T.N.C, Proudfoot, A.E.I. | | Deposit date: | 2004-07-26 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray structure of RANTES: heparin-derived disaccharides allows the rational design of chemokine inhibitors.
Structure, 12, 2004
|
|
1U4M
 
 | | human RANTES complexed to heparin-derived disaccharide III-S | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose, ACETIC ACID, Small inducible cytokine A5 | | Authors: | Shaw, J.P, Johnson, Z, Borlat, F, Zwahlen, C, Kungl, A, Roulin, K, Harrenga, A, Wells, T.N.C, Proudfoot, A.E.I. | | Deposit date: | 2004-07-26 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray structure of RANTES: heparin-derived disaccharides allows the rational design of chemokine inhibitors.
Structure, 12, 2004
|
|
1U4P
 
 | | Crystal Structure of human RANTES mutant K45E | | Descriptor: | ACETIC ACID, Small inducible cytokine A5 | | Authors: | Shaw, J.P, Johnson, Z, Borlat, F, Zwahlen, C, Kungl, A, Roulin, K, Harrenga, A, Wells, T.N.C, Proudfoot, A.E.I. | | Deposit date: | 2004-07-26 | | Release date: | 2004-11-09 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The X-ray structure of RANTES: heparin-derived disaccharides allows the rational design of chemokine inhibitors.
Structure, 12, 2004
|
|
7SF6
 
 | | Crystal Structure of Siderophore Binding Protein FatB from Desulfitobacterium hafniense | | Descriptor: | 1,2-ETHANEDIOL, 2-(2,3-DIHYDROXY-BENZOYLAMINO)-3-HYDROXY-PROPIONIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Patel, H.P, Nordquist, K.A, Schaab, K.M, Sha, J, Babnigg, G, Bond, A.H, Joachimiak, A, Midwest Center for Structural Genomics, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2021-10-03 | | Release date: | 2021-12-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal Structure of Siderophore Binding Protein FatB from Desulfitobacterium hafniense
To Be Published
|
|
