1AUA
 
 | | PHOSPHATIDYLINOSITOL TRANSFER PROTEIN SEC14P FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | PHOSPHATIDYLINOSITOL TRANSFER PROTEIN SEC14P, octyl beta-D-glucopyranoside | | Authors: | Sha, B, Phillips, S.E, Bankaitis, V.A, Luo, M. | | Deposit date: | 1997-08-20 | | Release date: | 1997-12-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol-transfer protein.
Nature, 391, 1998
|
|
1C3G
 
 | | S. CEREVISIAE HEAT SHOCK PROTEIN 40 SIS1 | | Descriptor: | HEAT SHOCK PROTEIN 40 | | Authors: | Sha, B, Lee, S, Cyr, D. | | Deposit date: | 1999-07-27 | | Release date: | 2000-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of the peptide-binding fragment from the yeast Hsp40 protein Sis1.
Structure, 8, 2000
|
|
1AA7
 
 | |
1HHO
 
 | | STRUCTURE OF HUMAN OXYHAEMOGLOBIN AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | HEMOGLOBIN A (OXY) (ALPHA CHAIN), HEMOGLOBIN A (OXY) (BETA CHAIN), OXYGEN MOLECULE, ... | | Authors: | Shaanan, B. | | Deposit date: | 1983-06-10 | | Release date: | 1983-10-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of human oxyhaemoglobin at 2.1 A resolution.
J.Mol.Biol., 171, 1983
|
|
5ZWT
 
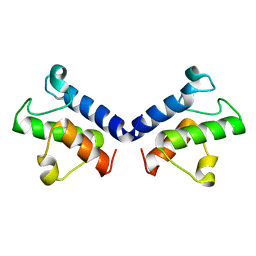 | |
7UG8
 
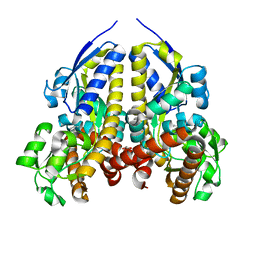 | | Crystal structure of a solute receptor from Synechococcus CC9311 in complex with alpha-ketovaleric and calcium | | Descriptor: | 1,2-ETHANEDIOL, 2-oxopentanoic acid, CALCIUM ION, ... | | Authors: | Shah, B.S, Mikolajek, H, Orr, C.M, Mykhaylyk, V, Owens, R.J, Paulsen, I.T. | | Deposit date: | 2022-03-24 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Crystal structure of a solute receptor from Synechococcus CC9311 in complex with alpha-ketovaleric and calcium
To Be Published
|
|
2V36
 
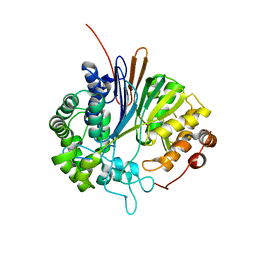 | | Crystal structure of gamma-glutamyl transferase from Bacillus subtilis | | Descriptor: | GAMMA-GLUTAMYLTRANSPEPTIDASE LARGE CHAIN, GAMMA-GLUTAMYLTRANSPEPTIDASE SMALL CHAIN | | Authors: | Sharath, B, Prabhune, A.A, Suresh, C.G, Wilkinson, A.J, Brannigan, J.A. | | Deposit date: | 2007-06-13 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Gamma-Glutamyl Transferase
To be Published
|
|
1LTE
 
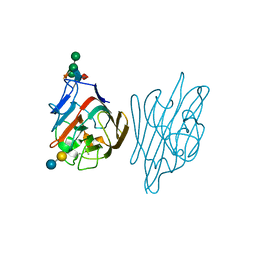 | | STRUCTURE OF A LEGUME LECTIN WITH AN ORDERED N-LINKED CARBOHYDRATE IN COMPLEX WITH LACTOSE | | Descriptor: | CALCIUM ION, CORAL TREE LECTIN, MANGANESE (II) ION, ... | | Authors: | Shaanan, B, Lis, H, Sharon, N. | | Deposit date: | 1991-06-25 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a legume lectin with an ordered N-linked carbohydrate in complex with lactose.
Science, 254, 1991
|
|
7S6F
 
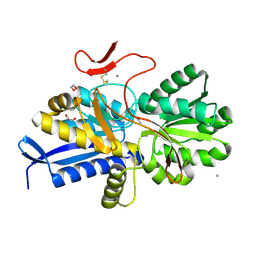 | | Crystal structure of UrtA1 from Synechococcus WH8102 in complex with urea and calcium | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Putative urea ABC transporter, ... | | Authors: | Shah, B.S, Mikolajek, H, Orr, C.M, Mykhaylyk, V, Owens, R.J, Paulsen, I.T. | | Deposit date: | 2021-09-14 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of UrtA1 from Synechococcus WH8102 in complex with urea and calcium
To Be Published
|
|
8BEO
 
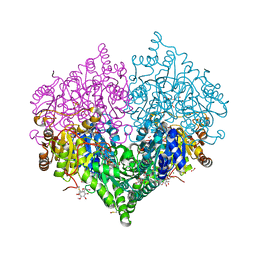 | | Crystal structure of E. coli glyoxylate carboligase mutant I393A with MAP | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2,3-DIMETHOXY-5-METHYL-1,4-BENZOQUINONE, ... | | Authors: | Shaanan, B, Binshtein, E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of E. coli glyoxylate carboligase mutant I393A with MAP
To Be Published
|
|
4XP8
 
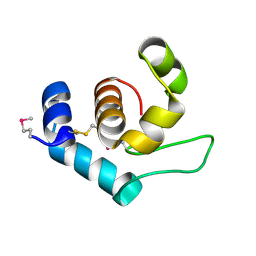 | |
7S6E
 
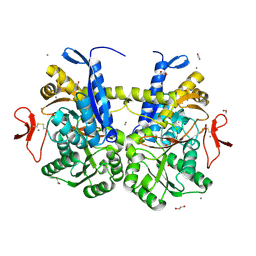 | | Crystal structure of UrtA from Synechococcus CC9311 in complex with urea and calcium | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Shah, B.S, Mikolajek, H, Mykhaylyk, V, Orr, C.M, Owens, R.J, Paulsen, I.T. | | Deposit date: | 2021-09-14 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Crystal structure of UrtA from Synechococcus CC9311 in complex with urea and calcium
To Be Published
|
|
6QUA
 
 | | The complex structure of hsRosR-SG (vng0258/RosR-SG) | | Descriptor: | DNA (28-MER), MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Shaanan, B, Kutnowski, N. | | Deposit date: | 2019-02-27 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.681 Å) | | Cite: | Specificity of protein-DNA interactions in hypersaline environment: structural studies on complexes of Halobacterium salinarum oxidative stress-dependent protein hsRosR.
Nucleic Acids Res., 47, 2019
|
|
1LIQ
 
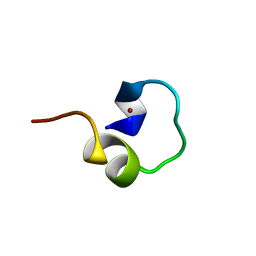 | | Non-native Solution Structure of a fragment of the CH1 domain of CBP | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Matthews, J.M, Kwan, A.H.Y, Newton, A, Gell, D.A, Crossley, M, Mackay, J.P. | | Deposit date: | 2002-04-18 | | Release date: | 2002-05-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A New Zinc Binding Fold Underlines the Versatility of Zinc Binding Modules in Protein Evolution
Structure, 10, 2002
|
|
4DID
 
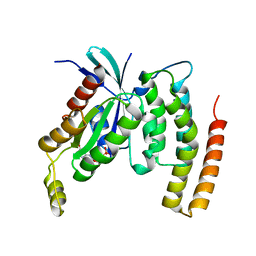 | |
1WO6
 
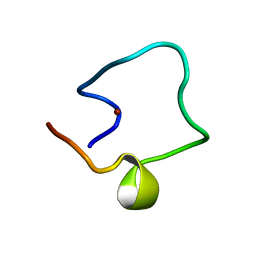 | | Solution structure of Designed Functional Finger 5 (DFF5): Designed mutant based on non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
1WO4
 
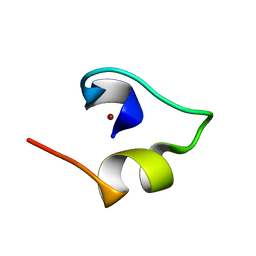 | | Solution structure of Minimal Mutant 2 (MM2): Multiple alanine mutant of non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
1WO5
 
 | | Solution structure of Designed Functional Finger 2 (DFF2): Designed mutant based on non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
1WO7
 
 | | Solution structure of Designed Functional Finger 7 (DFF7): Designed mutant based on non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
7S6G
 
 | | Crystal structure of PhnD from Synechococcus MITS9220 in complex with phosphate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Shah, B.S, Mikolajek, H, Orr, C.M, Mykhaylyk, V, Owens, R.J, Paulsen, I.T. | | Deposit date: | 2021-09-14 | | Release date: | 2021-10-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Marine picocyanobacterial PhnD1 shows specificity for various phosphorus sources but likely represents a constitutive inorganic phosphate transporter.
Isme J, 17, 2023
|
|
6QFD
 
 | | The complex structure of hsRosR-S4 (vng0258/RosR-S4) | | Descriptor: | DNA (28-MER), DNA-binding protein, MANGANESE (II) ION, ... | | Authors: | Shaanan, B, Kutnowski, N. | | Deposit date: | 2019-01-10 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.133 Å) | | Cite: | Specificity of protein-DNA interactions in hypersaline environment: structural studies on complexes of Halobacterium salinarum oxidative stress-dependent protein hsRosR.
Nucleic Acids Res., 47, 2019
|
|
6QH0
 
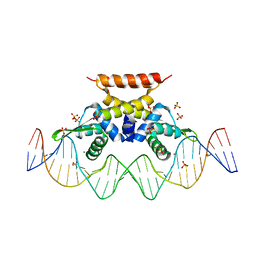 | | The complex structure of hsRosR-S5 (VNG0258H/RosR-S5) | | Descriptor: | DNA (28-MER), MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Shaanan, B, Kutnowski, N. | | Deposit date: | 2019-01-14 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.436 Å) | | Cite: | Specificity of protein-DNA interactions in hypersaline environment: structural studies on complexes of Halobacterium salinarum oxidative stress-dependent protein hsRosR.
Nucleic Acids Res., 47, 2019
|
|
6QIL
 
 | | The complex structure of hsRosR-S1 (VNG0258H/RosR-S1) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (28-MER), DNA binding protein, ... | | Authors: | Shaanan, B, Kutnowski, N. | | Deposit date: | 2019-01-21 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity of protein-DNA interactions in hypersaline environment: structural studies on complexes of Halobacterium salinarum oxidative stress-dependent protein hsRosR.
Nucleic Acids Res., 47, 2019
|
|
1WO3
 
 | | Solution structure of Minimal Mutant 1 (MM1): Multiple alanine mutant of non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
4J2O
 
 | | Crystal structure of NADP-bound WbjB from A. baumannii community strain D1279779 | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, UDP-N-acetylglucosamine 4,6-dehydratase/5-epimerase | | Authors: | Shah, B.S, Harrop, S.J, Paulsen, I.T, Mabbutt, B.C. | | Deposit date: | 2013-02-05 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Crystal structure of a UDP-GlcNAc epimerase for surface polysaccharide biosynthesis in Acinetobacter baumannii.
Plos One, 13, 2018
|
|
