3ZGG
 
 | | Crystal structure of the Fucosylgalactoside alpha N- acetylgalactosaminyltransferase (GTA, cisAB mutant L266G, G268A) in complex with NPE caged UDP-Gal (C222(1) space group) | | Descriptor: | 1-(2-NITROPHENYL)ETHYL UDP-GALACTOSE, GLYCEROL, HISTO-BLOOD GROUP ABO SYSTEM TRANSFERASE, ... | | Authors: | Jorgensen, R, Batot, G.O, Hindsgaul, O, Tanaka, H, Perez, S, Imberty, A, Breton, C, Royant, A, Palcic, M.M. | | Deposit date: | 2012-12-17 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of a Human Blood Group Glycosyltransferase in Complex with a Photo-Activatable Udp-Gal Derivative Reveal Two Different Binding Conformations
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3E65
 
 | | Murine INOS dimer with HEME, pterin and inhibitor AR-C120011 | | Descriptor: | 1-[4-(AMINOMETHYL)BENZOYL]-5'-FLUORO-1'H-SPIRO[PIPERIDINE-4,2'-QUINAZOLIN]-4'-AMINE, 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Rosenfeld, R.J, Garcin, E.D, Getzoff, E.D. | | Deposit date: | 2008-08-14 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
5JYU
 
 | |
5JYT
 
 | |
5JWO
 
 | | Crystal structure of foldswitch-stabilized KaiB in complex with the N-terminal CI domain of KaiC from Thermosynechococcus elongatus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Circadian clock protein KaiB, Circadian clock protein kinase KaiC | | Authors: | Tseng, R, Goularte, N.F, Chavan, A, Luu, J, Chang, Y, Heilser, J, Tripathi, S, LiWang, A, Partch, C.L. | | Deposit date: | 2016-05-12 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the day-night transition in a bacterial circadian clock.
Science, 355, 2017
|
|
3ZGF
 
 | | Crystal structure of the Fucosylgalactoside alpha N- acetylgalactosaminyltransferase (GTA, cisAB mutant L266G, G268A) in complex with in complex with NPE caged UDP-Gal (P2(1)2(1)2(1) space group) | | Descriptor: | 1-(2-NITROPHENYL)ETHYL UDP-GALACTOSE, HISTO-BLOOD GROUP ABO SYSTEM TRANSFERASE, MANGANESE (II) ION, ... | | Authors: | Jorgensen, R, Batot, G.O, Hindsgaul, O, Tanaka, H, Perez, S, Imberty, A, Breton, C, Royant, A, Palcic, M.M. | | Deposit date: | 2012-12-17 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structures of a Human Blood Group Glycosyltransferase in Complex with a Photo-Activatable Udp-Gal Derivative Reveal Two Different Binding Conformations
Acta Crystallogr.,Sect.F, 70, 2014
|
|
7P2D
 
 | | Structure of alphaMbeta2/Cd11bCD18 headpiece in complex with a nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Jensen, R.K, Andersen, G.R. | | Deposit date: | 2021-07-05 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the function-modulating effects of nanobody binding to the integrin receptor alpha M beta 2.
J.Biol.Chem., 298, 2022
|
|
6EHG
 
 | | complement component C3b in complex with a nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jensen, R.K, Andersen, K.R, Gadeberg, T.A.F, Laursen, N.S, Andersen, G.R. | | Deposit date: | 2017-09-13 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A potent complement factor C3-specific nanobody inhibiting multiple functions in the alternative pathway of human and murine complement.
J. Biol. Chem., 293, 2018
|
|
4X7S
 
 | |
4X7T
 
 | |
6YO6
 
 | | Structure of iC3b1 | | Descriptor: | hC3Nb1, iC3b1 alpha chain, iC3b1 beta chain | | Authors: | Jensen, R.K, Andersen, G.R. | | Deposit date: | 2020-04-14 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Complement Receptor 3 Forms a Compact High-Affinity Complex with iC3b.
J Immunol., 206, 2021
|
|
6RU5
 
 | | human complement C3 in complex with the hC3Nb1 nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jensen, R.K, Andersen, G.R. | | Deposit date: | 2019-05-27 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural Basis for Properdin Oligomerization and Convertase Stimulation in the Human Complement System.
Front Immunol, 10, 2019
|
|
8DHB
 
 | | Active FLCN GAP complex | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Folliculin, Folliculin-interacting protein 2, ... | | Authors: | Jansen, R.M, Hurley, J.H. | | Deposit date: | 2022-06-25 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural basis for FLCN RagC GAP activation in MiT-TFE substrate-selective mTORC1 regulation.
Sci Adv, 8, 2022
|
|
7AB5
 
 | |
7AB4
 
 | |
7AB3
 
 | |
6B0Y
 
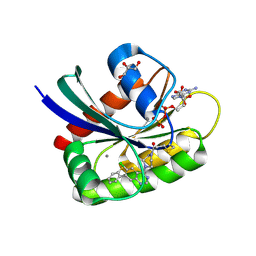 | | Crystal Structure of small molecule ARS-917 covalently bound to K-Ras G12C | | Descriptor: | 1-{4-[6-chloro-7-(2-fluorophenyl)quinazolin-4-yl]piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | Authors: | Hansen, R, Peters, U, Babbar, A, Chen, Y, Feng, J, Janes, M.R, Li, L.-S, Ren, P, Liu, Y, Zarrinkar, P.P. | | Deposit date: | 2017-09-15 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The reactivity-driven biochemical mechanism of covalent KRASG12Cinhibitors.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6B0V
 
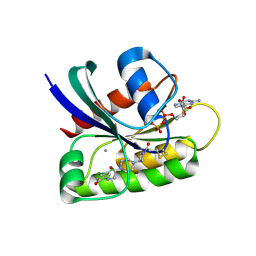 | | Crystal Structure of small molecule ARS-107 covalently bound to K-Ras G12C | | Descriptor: | 1-[3-(4-{[(4,5-dichloro-2-hydroxyphenyl)amino]acetyl}piperazin-1-yl)azetidin-1-yl]propan-1-one, CALCIUM ION, GTPase KRas, ... | | Authors: | Hansen, R, Peters, U, Babbar, A, Chen, Y, Feng, J, Janes, M.R, Li, L.-S, Ren, P, Liu, Y, Zarrinkar, P.P. | | Deposit date: | 2017-09-15 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | The reactivity-driven biochemical mechanism of covalent KRASG12Cinhibitors.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4X2C
 
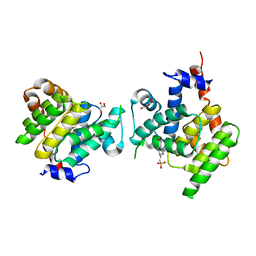 | | Clostridium difficile Fic protein_0569 mutant S31A, E35A | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fic family protein putative filamentation induced by cAMP protein, GLYCEROL, ... | | Authors: | Jorgensen, R, Dedic, E. | | Deposit date: | 2014-11-26 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Clostridium difficile Fic_0569 S31A, E35A mutant at 1.8 Angstroms resolution
To Be Published
|
|
7NP9
 
 | |
4X2E
 
 | | Clostridium difficile wild type Fic protein | | Descriptor: | Fic family protein putative filamentation induced by cAMP protein | | Authors: | Jorgensen, R, Dedic, E. | | Deposit date: | 2014-11-26 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | Structure of wild type Clostridium difficile Fic_0569 at 3.1 Angstroms resolution
To Be Published
|
|
4X2D
 
 | |
8DUT
 
 | | Duplex-G-quadruplex-duplex (DGD) class_1 | | Descriptor: | Duplex-G-quadruplex-duplex (46-MER) | | Authors: | Monsen, R.C, Chua, E.Y.D, Hopkins, J.B, Chaires, J.B, Trent, J.O. | | Deposit date: | 2022-07-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure of a 28.5 kDa duplex-embedded G-quadruplex system resolved to 7.4 angstrom resolution with cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
5JWR
 
 | | Crystal structure of foldswitch-stabilized KaiB in complex with the N-terminal CI domain of KaiC and a dimer of KaiA C-terminal domains from Thermosynechococcus elongatus | | Descriptor: | Circadian clock protein KaiA, Circadian clock protein KaiB, Circadian clock protein kinase KaiC, ... | | Authors: | Tseng, R, Goularte, N.F, Chavan, A, Luu, J, Chang, Y.G, Heilser, J, Tripathi, S, LiWang, A, Partch, C.L. | | Deposit date: | 2016-05-12 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis of the day-night transition in a bacterial circadian clock.
Science, 355, 2017
|
|
5JYV
 
 | |
