3QO3
 
 | | Crystal structure of Escherichia coli Hfq, in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Protein hfq | | Authors: | Beich-Frandsen, M, Vecerek, B, Hammele, H, Kloiber, K, Sjoeblom, B, Blasi, U, Djinovic-Carugo, K. | | Deposit date: | 2011-02-09 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and biochemical studies on ATP binding and hydrolysis by the Escherichia coli RNA chaperone Hfq
Plos One, 7, 2012
|
|
8B2E
 
 | | Muramidase from Kionochaeta sp natural catalytic core | | Descriptor: | CADMIUM ION, Muramidase | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Skov, L.K, Pache, R.A, Schnorr, K.M, Kiemer, L, Nymand-Grarup, S, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Davies, G.J, Wilson, K.S. | | Deposit date: | 2022-09-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Module walking using an SH3-like cell-wall-binding domain leads to a new GH184 family of muramidases.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8B2G
 
 | | SH3-like domain from Penicillium virgatum muramidase | | Descriptor: | 1,2-ETHANEDIOL, SH3b domain-containing protein, ZINC ION | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Skov, L.K, Pache, R.A, Schnorr, K.M, Kiemer, L, Nymand-Grarup, S, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Davies, G.J, Wilson, K.S. | | Deposit date: | 2022-09-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Module walking using an SH3-like cell-wall-binding domain leads to a new GH184 family of muramidases.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8B2H
 
 | | Muramidase from Thermothielavioides terrestris, catalytic domain | | Descriptor: | 1,2-ETHANEDIOL, SH3b domain-containing protein, ZINC ION | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Skov, L.K, Pache, R.A, Schnorr, K.M, Kiemer, L, Nymand-Grarup, S, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Davies, G.J, Wilson, K.S. | | Deposit date: | 2022-09-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Module walking using an SH3-like cell-wall-binding domain leads to a new GH184 family of muramidases.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8B2F
 
 | | SH3-like cell wall binding domain of the GH24 family muramidase from Trichophaea saccata in complex with triglycine | | Descriptor: | 1,2-ETHANEDIOL, GLY-GLY-GLY, SH3-like cell wall binding domain-containing protein, ... | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Skov, L.K, Pache, R.A, Schnorr, K.M, Kiemer, L, Nymand-Grarup, S, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Davies, G.J, Wilson, K.S. | | Deposit date: | 2022-09-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.183 Å) | | Cite: | Module walking using an SH3-like cell-wall-binding domain leads to a new GH184 family of muramidases.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8B2S
 
 | | GH24 family muramidase from Trichophaea saccata with an SH3-like cell wall binding domain | | Descriptor: | GH24 family muramidase, POTASSIUM ION | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Skov, L.K, Pache, R.A, Schnorr, K.M, Kiemer, L, Nymand-Grarup, S, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Davies, G.J, Wilson, K.S. | | Deposit date: | 2022-09-14 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Module walking using an SH3-like cell-wall-binding domain leads to a new GH184 family of muramidases.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5YBL
 
 | | Fe(II)/(alpha)ketoglutarate-dependent dioxygenase AusE | | Descriptor: | 2-OXOGLUTARIC ACID, MANGANESE (II) ION, Multifunctional dioxygenase ausE | | Authors: | Nakashima, Y, Senda, M. | | Deposit date: | 2017-09-05 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | Structure function and engineering of multifunctional non-heme iron dependent oxygenases in fungal meroterpenoid biosynthesis.
Nat Commun, 9, 2018
|
|
5IPI
 
 | | Structure of Adeno-associated virus type 2 VLP | | Descriptor: | Capsid protein VP1 | | Authors: | Drouin, L.M, Lins, B, Janssen, M.E, Bennet, A, Chipman, P, McKenna, R, Chen, W, Muzyczka, N, Cardone, G, Baker, T.S, Agbandje-McKenna, M. | | Deposit date: | 2016-03-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-electron Microscopy Reconstruction and Stability Studies of the Wild Type and the R432A Variant of Adeno-associated Virus Type 2 Reveal that Capsid Structural Stability Is a Major Factor in Genome Packaging.
J.Virol., 90, 2016
|
|
7LTE
 
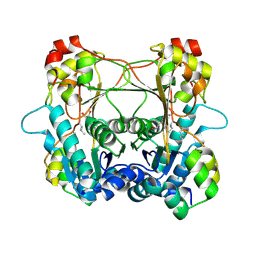 | | Structure of the alpha-N-methyltransferase (SonM) and RiPP precursor (SonA) heteromeric complex (with SAH) | | Descriptor: | LigA domain-containing protein, S-ADENOSYL-L-HOMOCYSTEINE, TP-methylase family protein | | Authors: | Miller, F.S, Crone, K.K, Jensen, M.R, Shaw, S, Harcombe, W.R, Elias, M, Freeman, M.F. | | Deposit date: | 2021-02-19 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational rearrangements enable iterative backbone N-methylation in RiPP biosynthesis.
Nat Commun, 12, 2021
|
|
7LTC
 
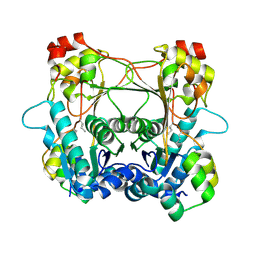 | | Structure of the alpha-N-methyltransferase (SonM) and RiPP precursor (SonA) heteromeric complex (no cofactor) | | Descriptor: | LigA domain-containing protein, TP-methylase family protein | | Authors: | Miller, F.S, Crone, K.K, Jensen, M.R, Shaw, S, Harcombe, W.R, Elias, M, Freeman, M.F. | | Deposit date: | 2021-02-19 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational rearrangements enable iterative backbone N-methylation in RiPP biosynthesis.
Nat Commun, 12, 2021
|
|
7LTF
 
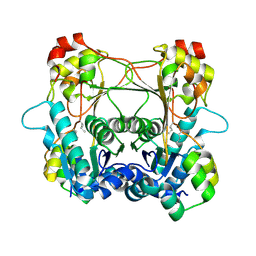 | | Structure of the alpha-N-methyltransferase (SonM mutant Y58F) and RiPP precursor (SonA) heteromeric complex (no cofactor) | | Descriptor: | LigA domain-containing protein, TP-methylase family protein | | Authors: | Miller, F.S, Crone, K.K, Jensen, M.R, Shaw, S, Harcombe, W.R, Elias, M, Freeman, M.F. | | Deposit date: | 2021-02-19 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational rearrangements enable iterative backbone N-methylation in RiPP biosynthesis.
Nat Commun, 12, 2021
|
|
7LTR
 
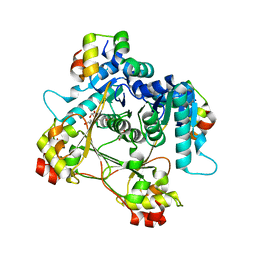 | | Structure of the heteromeric complex between the alpha-N-methyltransferase (SonM) and a truncated construct of the RiPP precursor (SonA) (with SAM) | | Descriptor: | GLYCEROL, LigA domain-containing protein, S-ADENOSYLMETHIONINE, ... | | Authors: | Miller, F.S, Crone, K.K, Jensen, M.R, Shaw, S, Harcombe, W.R, Elias, M, Freeman, M.F. | | Deposit date: | 2021-02-19 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Conformational rearrangements enable iterative backbone N-methylation in RiPP biosynthesis.
Nat Commun, 12, 2021
|
|
7LTH
 
 | | Structure of the alpha-N-methyltransferase (SonM mutant Y93F) and RiPP precursor (SonA) heteromeric complex (no cofactor) | | Descriptor: | LigA domain-containing protein, TP-methylase family protein | | Authors: | Miller, F.S, Crone, K.K, Jensen, M.R, Shaw, S, Harcombe, W.R, Elias, M, Freeman, M.F. | | Deposit date: | 2021-02-19 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational rearrangements enable iterative backbone N-methylation in RiPP biosynthesis.
Nat Commun, 12, 2021
|
|
7LTS
 
 | | Structure of the alpha-N-methyltransferase (SonM mutant R67A) and RiPP precursor (SonA) heteromeric complex (with SAH) | | Descriptor: | LigA domain-containing protein, S-ADENOSYL-L-HOMOCYSTEINE, TP-methylase family protein | | Authors: | Miller, F.S, Crone, K.K, Jensen, M.R, Shaw, S, Harcombe, W.R, Elias, M, Freeman, M.F. | | Deposit date: | 2021-02-20 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Conformational rearrangements enable iterative backbone N-methylation in RiPP biosynthesis.
Nat Commun, 12, 2021
|
|
5IPK
 
 | | Structure of the R432A variant of Adeno-associated virus type 2 VLP | | Descriptor: | Capsid protein VP1 | | Authors: | Drouin, L.M, Lins, B, Janssen, M.E, Bennet, A, Chipman, P, McKenna, R, Chen, W, Muzyczka, N, Cardone, G, Baker, T.S, Agbandje-McKenna, M. | | Deposit date: | 2016-03-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-electron Microscopy Reconstruction and Stability Studies of the Wild Type and the R432A Variant of Adeno-associated Virus Type 2 Reveal that Capsid Structural Stability Is a Major Factor in Genome Packaging.
J.Virol., 90, 2016
|
|
7AJ0
 
 | | Crystal structure of PsFucS1 sulfatase from Pseudoalteromonas sp. | | Descriptor: | Arylsulfatase, CALCIUM ION, CHLORIDE ION | | Authors: | Roret, T, Mikkelsen, M.D, Czjzek, M, Meyer, A.S. | | Deposit date: | 2020-09-28 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A novel thermostable prokaryotic fucoidan active sulfatase PsFucS1 with an unusual quaternary hexameric structure.
Sci Rep, 11, 2021
|
|
5FYF
 
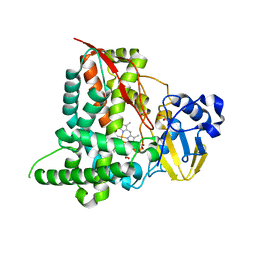 | | Structure of CYP153A from Marinobacter aquaeolei | | Descriptor: | 1,2-ETHANEDIOL, CYTOCHROME P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Danesh Azari, H.R, Spandolf, C, Hoffman, S.M, Weissenborn, M, Hauer, B, Grogan, G. | | Deposit date: | 2016-03-07 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure-Guided Redesign of CYP153AM.aqfor the Improved Terminal Hydroxylation of Fatty Acids
Chemcatchem, 2016
|
|
7M2G
 
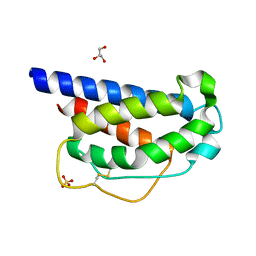 | | INTERLEUKIN-2 (human) mutant P65K, C125S | | Descriptor: | GLYCEROL, Interleukin-2, SULFATE ION | | Authors: | Ptacin, J.L, Milla, M.E, Steinbacher, S, Maskos, K, Thomsen, M. | | Deposit date: | 2021-03-16 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | An engineered IL-2 reprogrammed for anti-tumor therapy using a semi-synthetic organism.
Nat Commun, 12, 2021
|
|
3U8L
 
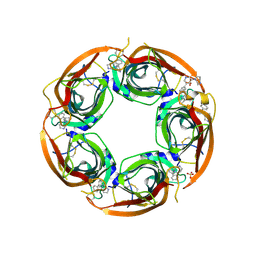 | | Crystal structure of the acetylcholine binding protein (AChBP) from Lymnaea stagnalis in complex with NS3570 (1-(5-phenylpyridin-3-yl)-1,4-diazepane) | | Descriptor: | 1-(5-phenylpyridin-3-yl)-1,4-diazepane, Acetylcholine-binding protein, SULFATE ION | | Authors: | Rohde, L.A.H, Ahring, P.K, Jensen, M.L, Nielsen, E.O, Peters, D, Helgstrand, C, Krintel, C, Harpsoe, K, Gajhede, M, Kastrup, J.S, Balle, T. | | Deposit date: | 2011-10-17 | | Release date: | 2011-12-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Intersubunit bridge formation governs agonist efficacy at nicotinic acetylcholine alpha 4 beta 2 receptors: unique role of halogen bonding revealed.
J.Biol.Chem., 287, 2012
|
|
3QHS
 
 | | Crystal structure of full-length Hfq from Escherichia coli | | Descriptor: | Protein hfq | | Authors: | Beich-Frandsen, M, Vecerek, B, Sjoeblom, B, Blaesi, U, Djinovic-Carugo, K. | | Deposit date: | 2011-01-26 | | Release date: | 2011-05-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural analysis of full-length Hfq from Escherichia coli
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3U8J
 
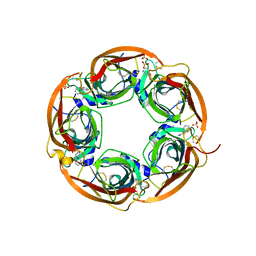 | | Crystal structure of the acetylcholine binding protein (AChBP) from Lymnaea stagnalis in complex with NS3531 (1-(pyridin-3-yl)-1,4-diazepane) | | Descriptor: | 1-(pyridin-3-yl)-1,4-diazepane, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, ... | | Authors: | Rohde, L.A.H, Ahring, P.K, Jensen, M.L, Nielsen, E.O, Peters, D, Helgstrand, C, Krintel, C, Harpsoe, K, Gajhede, M, Kastrup, J.S, Balle, T. | | Deposit date: | 2011-10-17 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Intersubunit bridge formation governs agonist efficacy at nicotinic acetylcholine alpha 4 beta 2 receptors: unique role of halogen bonding revealed.
J.Biol.Chem., 287, 2012
|
|
8DAD
 
 | | SARS-CoV-2 receptor binding domain in complex with AZ090 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AZ090 Fab Heavy Chain, AZ090 Fab Light Chain, ... | | Authors: | Zong, S, Wang, Z, Gaebler, C, Nussenzweig, M. | | Deposit date: | 2022-06-13 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | SARS-CoV-2 receptor binding domain in complex with AZ090 Fab
To Be Published
|
|
6H2B
 
 | | Structure of the Macrobrachium rosenbergii Nodavirus | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Ho, K.H, Gabrielsen, M, Beh, P.L, Kueh, C.L, Thong, Q.X, Streetley, J, Tan, W.S, Bhella, D. | | Deposit date: | 2018-07-13 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure of the Macrobrachium rosenbergii nodavirus: A new genus within the Nodaviridae?
PLoS Biol., 16, 2018
|
|
7ZQI
 
 | | MHC class I from a wild bird in complex with a nonameric peptide P2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Eltschkner, S, Mellinger, S, Buus, S, Nielsen, M, Paulsson, K.M, Lindkvist-Petersson, K, Westerdahl, H. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of songbird MHC class I reveals antigen binding that is flexible at the N-terminus and static at the C-terminus.
Front Immunol, 14, 2023
|
|
7ZQJ
 
 | | MHC class I from a wild bird in complex with a nonameric peptide P3 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, MHC class I antigen, ... | | Authors: | Eltschkner, S, Mellinger, S, Buus, S, Nielsen, M, Paulsson, K.M, Lindkvist-Petersson, K, Westerdahl, H. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure of songbird MHC class I reveals antigen binding that is flexible at the N-terminus and static at the C-terminus.
Front Immunol, 14, 2023
|
|
