5L51
 
 | |
2O2K
 
 | | Crystal Structure of the Activation Domain of Human Methionine Synthase Isoform/Mutant D963E/K1071N | | Descriptor: | Methionine synthase | | Authors: | Wolthers, K.R, Toogood, H.S, Jowitt, T.A, Marshall, K.R, Leys, D, Scrutton, N.S. | | Deposit date: | 2006-11-30 | | Release date: | 2006-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure and solution characterization of the activation domain of human methionine synthase
Febs J., 274, 2007
|
|
2QTL
 
 | | Crystal Structure of the FAD-containing FNR-like Module of Human Methionine Synthase Reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Methionine synthase reductase | | Authors: | Wolthers, K.R, Lou, X, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2007-08-02 | | Release date: | 2007-11-13 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of Coenzyme Binding to Human Methionine Synthase Reductase Revealed through the Crystal Structure of the FNR-like Module and Isothermal Titration Calorimetry
Biochemistry, 46, 2007
|
|
2QTZ
 
 | | Crystal Structure of the NADP+-bound FAD-containing FNR-like Module of Human Methionine Synthase Reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Methionine synthase reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wolthers, K.R, Lou, X, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2007-08-02 | | Release date: | 2007-11-13 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of Coenzyme Binding to Human Methionine Synthase Reductase Revealed through the Crystal Structure of the FNR-like Module and Isothermal Titration Calorimetry
Biochemistry, 46, 2007
|
|
2R14
 
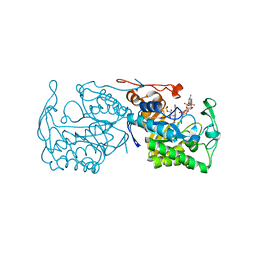 | | Structure of morphinone reductase in complex with tetrahydroNAD | | Descriptor: | 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN MONONUCLEOTIDE, Morphinone reductase | | Authors: | Costello, C.L, Scrutton, N.S, Leys, D. | | Deposit date: | 2007-08-22 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mutagenesis of morphinone reductase induces multiple reactive configurations and identifies potential ambiguity in kinetic analysis of enzyme tunneling mechanisms.
J.Am.Chem.Soc., 129, 2007
|
|
2XX1
 
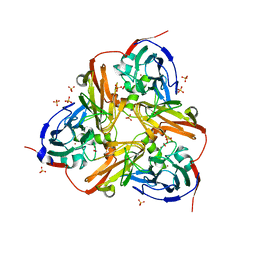 | | STRUCTURE OF THE N90S MUTANT OF NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS complexed with nitrite | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, NITRITE ION, ... | | Authors: | Antonyuk, S.V, Leferink, N.G.H, Han, C, Heyes, D.J, Rigby, S.E.J, Hough, M.A, Eady, R.R, Scrutton, N.S, Hasnain, S.S. | | Deposit date: | 2010-11-07 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Proton-Coupled Electron Transfer in the Catalytic Cycle of Alcaligenes Xylosoxidans Copper-Dependent Nitrite Reductase.
Biochemistry, 50, 2011
|
|
2XX0
 
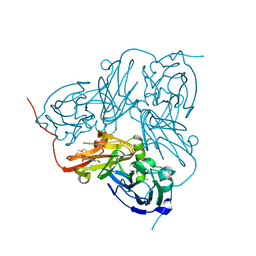 | | STRUCTURE OF THE N90S-H254F MUTANT OF NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Antonyuk, S.V, Leferink, N.G.H, Han, C, Heyes, D.J, Rigby, S.E.J, Hough, M.A, Eady, R.R, Scrutton, N.S, Hasnain, S.S. | | Deposit date: | 2010-11-07 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Proton-Coupled Electron Transfer in the Catalytic Cycle of Alcaligenes Xylosoxidans Copper-Dependent Nitrite Reductase.
Biochemistry, 50, 2011
|
|
2XWZ
 
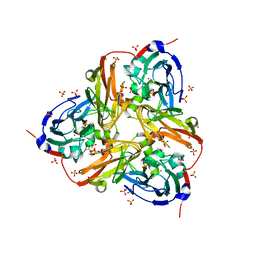 | | STRUCTURE OF THE RECOMBINANT NATIVE NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS complexed with nitrite | | Descriptor: | ACETATE ION, COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Antonyuk, S.V, Leferink, N.G.H, Han, C, Heyes, D.J, Rigby, S.E.J, Hough, M.A, Eady, R.R, Scrutton, N.S, Hasnain, S.S. | | Deposit date: | 2010-11-06 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Proton-Coupled Electron Transfer in the Catalytic Cycle of Alcaligenes Xylosoxidans Copper-Dependent Nitrite Reductase.
Biochemistry, 50, 2011
|
|
6ER9
 
 | |
3P62
 
 | |
3P80
 
 | |
3P8I
 
 | | Y351F mutant of pentaerythritol tetranitrate reductase containing a bound acetate molecule | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Pentaerythritol tetranitrate reductase | | Authors: | Toogood, H.S, Scrutton, N.S. | | Deposit date: | 2010-10-14 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Active site modifications in pentaerythritol tetranitrate reductase can lead to improved product enantiopurity, decreased by-product formation and altered stereochemical outcome in reactions with a,b-unsaturated nitroolefins
Catalysis Science and Technology, 2011
|
|
3P82
 
 | |
3P7Y
 
 | |
3P81
 
 | |
3P67
 
 | |
3P74
 
 | |
3P8J
 
 | |
3P84
 
 | | Y351A mutant of pentaerythritol tetranitrate reductase containing a bound acetate molecule | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Pentaerythritol tetranitrate reductase | | Authors: | Toogood, H.S, Scrutton, N.S. | | Deposit date: | 2010-10-13 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Active site modifications in pentaerythritol tetranitrate reductase can lead to improved product enantiopurity, decreased by-product formation and altered stereochemical outcome in reactions with a,b-unsaturated nitroolefins
Catalysis Science and Technology, 2011
|
|
4HFN
 
 | |
4J31
 
 | | Crystal Structure of kynurenine 3-monooxygenase (KMO-396Prot) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Amaral, M, Levy, C, Heyes, D.J, Lafite, P, Outeiro, T.F, Giorgini, F, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-04-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of kynurenine 3-monooxygenase inhibition.
Nature, 496, 2013
|
|
4J2W
 
 | | Crystal Structure of kynurenine 3-monooxygenase (KMO-396Prot-Se) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Amaral, M, Levy, C, Heyes, D.J, Lafite, P, Outeiro, T.F, Giorgini, F, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of kynurenine 3-monooxygenase inhibition.
Nature, 496, 2013
|
|
4HFM
 
 | |
4GU5
 
 | | Structure of Full-length Drosophila Cryptochrome | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION | | Authors: | Zoltowski, B.D, Vaidya, A.T, Top, D, Widom, J, Young, M.W, Levy, C, Jones, A.R, Scrutton, N.S, Leys, D, Crane, B.R. | | Deposit date: | 2012-08-29 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Updated structure of Drosophila cryptochrome.
Nature, 495, 2013
|
|
4HFJ
 
 | |
