2LYP
 
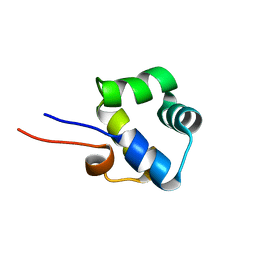 | | NOE-based 3D structure of the monomer of CylR2 in equilibrium with predissociated homodimer at 266K (-7 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LWA
 
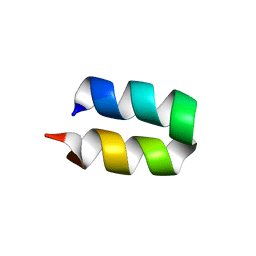 | | Conformational ensemble for the G8A mutant of the influenza hemagglutinin fusion peptide | | Descriptor: | HEMAGGLUTININ FUSION PEPTIDE G8A MUTANT | | Authors: | Lorieau, J.L, Louis, J.M, Schwieters, C.D, Bax, A. | | Deposit date: | 2012-07-26 | | Release date: | 2012-12-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | pH-triggered, activated-state conformations of the influenza hemagglutinin fusion peptide revealed by NMR.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LYJ
 
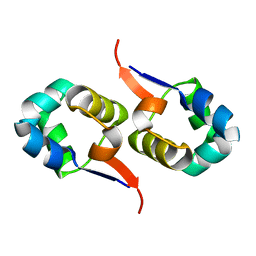 | | NOE-based 3D structure of the CylR2 homodimer at 298K | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Giller, K, Becker, S, Zweckstetter, M, Schwieters, C.D. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYK
 
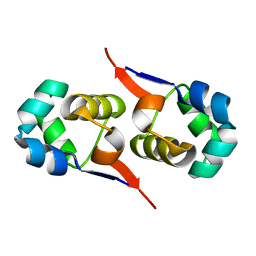 | | NOE-based 3D structure of the CylR2 homodimer at 270K (-3 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2M8P
 
 | | The structure of the W184AM185A mutant of the HIV-1 capsid protein | | Descriptor: | Capsid protein p24 | | Authors: | Deshmukh, L, Schwieters, C.D, Grishaev, A, Clore, G, Ghirlando, R. | | Deposit date: | 2013-05-24 | | Release date: | 2013-11-20 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Structure and Dynamics of Full-Length HIV-1 Capsid Protein in Solution.
J.Am.Chem.Soc., 135, 2013
|
|
2M02
 
 | | 3D structure of cap-gly domain of mammalian dynactin determined by magic angle spinning NMR spectroscopy | | Descriptor: | Dynactin subunit 1 | | Authors: | Yan, S, Hou, G, Schwieters, C.D, Ahmed, S, Williams, J.C, Polenova, T. | | Deposit date: | 2012-10-15 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Three-Dimensional Structure of CAP-Gly Domain of Mammalian Dynactin Determined by Magic Angle Spinning NMR Spectroscopy: Conformational Plasticity and Interactions with End-Binding Protein EB1.
J.Mol.Biol., 425, 2013
|
|
2M8N
 
 | | HIV-1 capsid monomer structure | | Descriptor: | Capsid protein p24 | | Authors: | Deshmukh, L, Schwieters, C.D, Grishaev, A, Clore, G, Ghirlando, R. | | Deposit date: | 2013-05-24 | | Release date: | 2013-11-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Structure and Dynamics of Full-Length HIV-1 Capsid Protein in Solution.
J.Am.Chem.Soc., 135, 2013
|
|
2N9Z
 
 | | Solution structure of K1 lobe of double-knot toxin | | Descriptor: | Tau-theraphotoxin-Hs1a | | Authors: | Bae, C, Anselmi, C, Kalia, J, Jara-Oseguera, A, Schwieters, C.D, Krepkiy, D, Lee, C.W, Kim, E.H, Kim, J.I, Faraldo-Gomez, J.D, Swartz, K.J. | | Deposit date: | 2015-12-16 | | Release date: | 2016-03-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the mechanism of activation of the TRPV1 channel by a membrane-bound tarantula toxin
Elife, 5, 2016
|
|
2NAJ
 
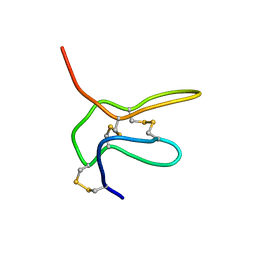 | | Solution structure of K2 lobe of double-knot toxin | | Descriptor: | Tau-theraphotoxin-Hs1a | | Authors: | Bae, C, Anselmi, C, Kalia, J, Jara-Oseguera, A, Schwieters, C.D, Krepkiy, D, Lee, C.W, Kim, E.H, Kim, J.I, Faraldo-Gomez, J.D, Swartz, K.J. | | Deposit date: | 2016-01-04 | | Release date: | 2016-03-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the mechanism of activation of the TRPV1 channel by a membrane-bound tarantula toxin
Elife, 5, 2016
|
|
2N5T
 
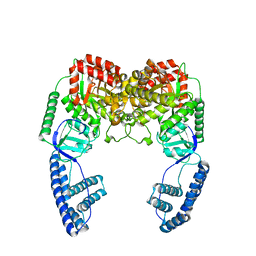 | | Ensemble solution structure of the phosphoenolpyruvate-Enzyme I complex from the bacterial phosphotransferase system | | Descriptor: | Phosphoenolpyruvate-protein phosphotransferase | | Authors: | Venditti, V, Schwieters, C.D, Grishaev, A, Clore, G. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Dynamic equilibrium between closed and partially closed states of the bacterial Enzyme I unveiled by solution NMR and X-ray scattering.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2LEG
 
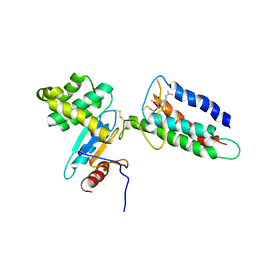 | | Membrane protein complex DsbB-DsbA structure by joint calculations with solid-state NMR and X-ray experimental data | | Descriptor: | Disulfide bond formation protein B, Thiol:disulfide interchange protein DsbA, UBIQUINONE-1, ... | | Authors: | Tang, M, Sperling, L.J, Berthold, D.A, Schwieters, C.D, Nesbitt, A.E, Nieuwkoop, A.J, Gennis, R.B, Rienstra, C.M. | | Deposit date: | 2011-06-15 | | Release date: | 2011-10-26 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution membrane protein structure by joint calculations with solid-state NMR and X-ray experimental data.
J.Biomol.Nmr, 51, 2011
|
|
2LYL
 
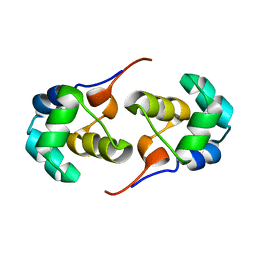 | | NOE-based 3D structure of the predissociated homodimer of CylR2 in equilibrium with monomer at 266K (-7 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYS
 
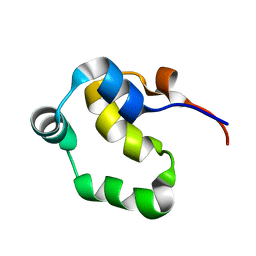 | | NOE-based 3D structure of the monomeric partially-folded intermediate of CylR2 at 257K (-16 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2N0A
 
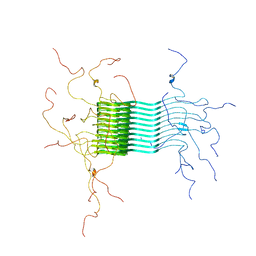 | | Atomic-resolution structure of alpha-synuclein fibrils | | Descriptor: | Alpha-synuclein | | Authors: | Tuttle, M.D, Comellas, G, Nieuwkoop, A.J, Covell, D.J, Berthold, D.A, Kloepper, K.D, Courtney, J.M, Kim, J.K, Schwieters, C.D, Lee, V.M, George, J.M, Rienstra, C.M. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Solid-state NMR structure of a pathogenic fibril of full-length human alpha-synuclein.
Nat.Struct.Mol.Biol., 23, 2016
|
|
2N2L
 
 | | NMR structure of yersinia pestis ail (attachment invasion locus) in decylphosphocholine micelles calculated with implicit membrane solvation | | Descriptor: | Outer membrane protein X | | Authors: | Marassi, F.M, Ding, Y, Tian, Y, Schwieters, C.D, Yao, Y. | | Deposit date: | 2015-05-10 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Backbone structure of Yersinia pestis Ail determined in micelles by NMR-restrained simulated annealing with implicit membrane solvation.
J.Biomol.Nmr, 63, 2015
|
|
2P01
 
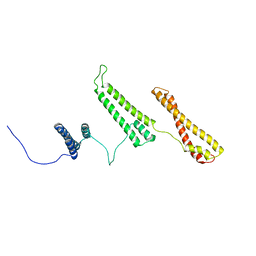 | | The structure of receptor-associated protein(RAP) | | Descriptor: | Alpha-2-macroglobulin receptor-associated protein | | Authors: | Lee, D, Walsh, J.D, Migliorini, M, Yu, P, Cai, T, Schwieters, C.D, Krueger, S, Strickland, D.K, Wang, Y.X. | | Deposit date: | 2007-02-28 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of receptor-associated protein (RAP).
Protein Sci., 16, 2007
|
|
2P03
 
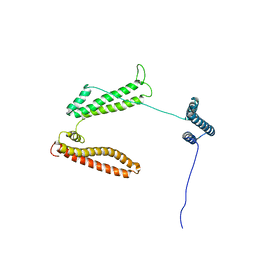 | | The structure of receptor-associated protein(RAP) | | Descriptor: | Alpha-2-macroglobulin receptor-associated protein | | Authors: | Lee, D, Walsh, J.D, Migliorini, M, Yu, P, Cai, T, Schwieters, C.D, Krueger, S, Strickland, D.K, Wang, Y.X. | | Deposit date: | 2007-02-28 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of receptor-associated protein (RAP).
Protein Sci., 16, 2007
|
|
2N74
 
 | | Solution Structure of the RNA-Binding domain of non-structural protein 1 from the 1918 H1N1 influenza virus | | Descriptor: | Non-structural protein 1 | | Authors: | Jureka, A.S, Kleinpeter, A.B, Cornilescu, G, Cornilescu, C.C, Schwieters, C.D, Petit, C.M. | | Deposit date: | 2015-09-03 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for a Novel Interaction between the NS1 Protein Derived from the 1918 Influenza Virus and RIG-I.
Structure, 23, 2015
|
|
