4PT7
 
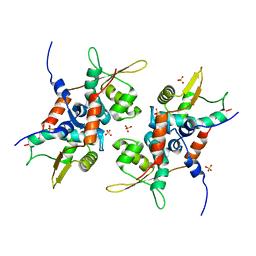 | | Structure of initiator | | Descriptor: | Replication initiator A family protein, SULFATE ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2014-03-10 | | Release date: | 2014-06-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of staphylococcal multiresistance plasmid replication origin assembly by the RepA protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4PTA
 
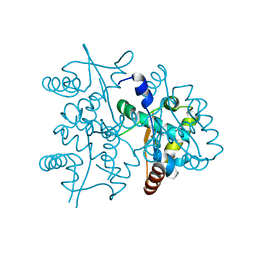 | | Structure of MDR initiator | | Descriptor: | Replication initiator protein | | Authors: | Schumacher, M.A. | | Deposit date: | 2014-03-10 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6003 Å) | | Cite: | Mechanism of staphylococcal multiresistance plasmid replication origin assembly by the RepA protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7TEA
 
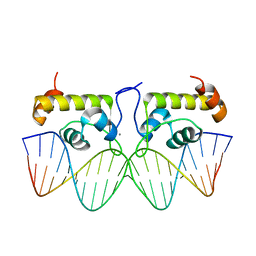 | | Crystal structure of S. aureus GlnR-DNA complex | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*GP*TP*GP*TP*CP*AP*GP*AP*TP*AP*AP*TP*CP*TP*GP*AP*CP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*GP*TP*CP*AP*GP*AP*TP*TP*AP*TP*CP*TP*GP*AP*CP*AP*CP*G)-3'), ... | | Authors: | Schumacher, M.A. | | Deposit date: | 2022-01-04 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
7TDP
 
 | |
7TDV
 
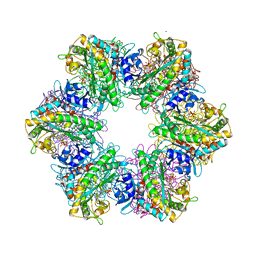 | |
7TEC
 
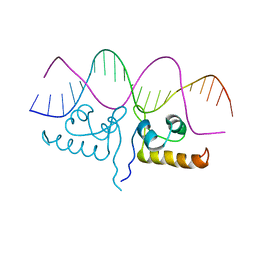 | |
7TEN
 
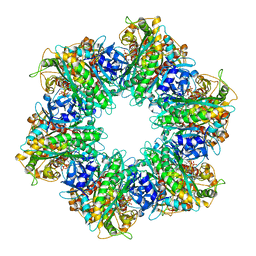 | |
7TZV
 
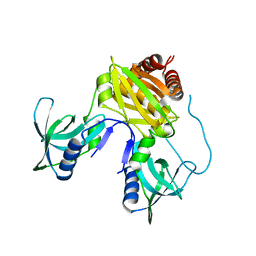 | | Structure of DriD C-domain bound to 9mer ssDNA | | Descriptor: | DNA (5'-D(*TP*AP*GP*TP*CP*TP*AP*CP*T)-3'), WYL domain-containing protein | | Authors: | Schumacher, M.A, Laub, M. | | Deposit date: | 2022-02-16 | | Release date: | 2022-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | ssDNA is an allosteric regulator of the C. crescentus SOS-independent DNA damage response transcription activator, DriD.
Genes Dev., 36, 2022
|
|
7U02
 
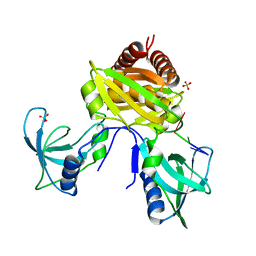 | | Structure of the C. crescentus DriD C-domain bound to ssDNA | | Descriptor: | DNA (5'-D(P*AP*CP*G)-3'), SULFATE ION, WYL domain-containing protein | | Authors: | Schumacher, M.A. | | Deposit date: | 2022-02-17 | | Release date: | 2022-06-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | ssDNA is an allosteric regulator of the C. crescentus SOS-independent DNA damage response transcription activator, DriD.
Genes Dev., 36, 2022
|
|
2G66
 
 | |
2GIA
 
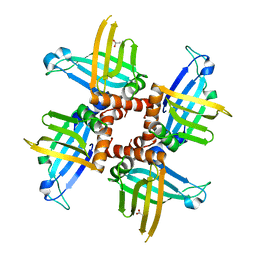 | | Crystal structures of trypanosoma bruciei MRP1/MRP2 | | Descriptor: | ACETIC ACID, mitochondrial RNA-binding protein 1, mitochondrial RNA-binding protein 2 | | Authors: | Schumacher, M.A, Karamooz, E, Zikova, A, Trantirek, L, Lukes, J. | | Deposit date: | 2006-03-28 | | Release date: | 2006-09-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structures of T. brucei MRP1/MRP2 Guide-RNA Binding Complex Reveal RNA Matchmaking Mechanism.
Cell(Cambridge,Mass.), 126, 2006
|
|
2GJE
 
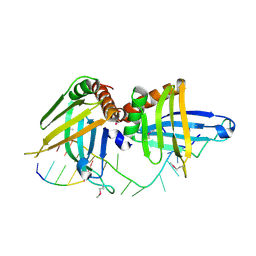 | | Structure of a guideRNA-binding protein complex bound to a gRNA | | Descriptor: | RNA tetramer, guide RNA 40-mer, mitochondrial RNA-binding protein 1, ... | | Authors: | Schumacher, M.A, Karamooz, E, Zikova, A, Trantirek, L, Lukes, J. | | Deposit date: | 2006-03-30 | | Release date: | 2006-09-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Crystal Structures of T. brucei MRP1/MRP2 Guide-RNA Binding Complex Reveal RNA Matchmaking Mechanism.
Cell(Cambridge,Mass.), 126, 2006
|
|
1DBQ
 
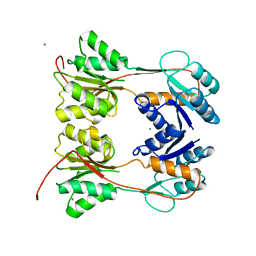 | | DNA-BINDING REGULATORY PROTEIN | | Descriptor: | MAGNESIUM ION, PURINE REPRESSOR | | Authors: | Schumacher, M.A, Choi, K.Y, Lu, F, Zalkin, H, Brennan, R.G. | | Deposit date: | 1996-02-13 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of corepressor-mediated specific DNA binding by the purine repressor.
Cell(Cambridge,Mass.), 83, 1995
|
|
2GID
 
 | | Crystal structures of trypanosoma bruciei MRP1/MRP2 | | Descriptor: | mitochondrial RNA-binding protein 1, mitochondrial RNA-binding protein 2 | | Authors: | Schumacher, M.A, Karamooz, E, Zikova, A, Trantirek, L, Lukes, J. | | Deposit date: | 2006-03-28 | | Release date: | 2006-09-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal Structures of T. brucei MRP1/MRP2 Guide-RNA Binding Complex Reveal RNA Matchmaking Mechanism.
Cell(Cambridge,Mass.), 126, 2006
|
|
4LNK
 
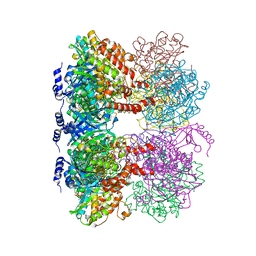 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: structure of GS-glutamate-AMPPCP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLUTAMIC ACID, Glutamine synthetase, ... | | Authors: | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | Deposit date: | 2013-07-11 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
4LNF
 
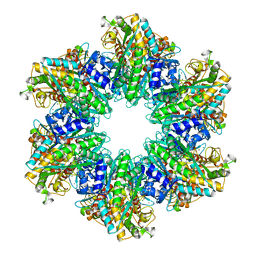 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: structure of GS-Q | | Descriptor: | GLUTAMINE, Glutamine synthetase, MAGNESIUM ION, ... | | Authors: | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | Deposit date: | 2013-07-11 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.949 Å) | | Cite: | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
4LNN
 
 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: structure of apo form of GS | | Descriptor: | Glutamine synthetase, MAGNESIUM ION, SULFATE ION | | Authors: | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | Deposit date: | 2013-07-11 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
4LNO
 
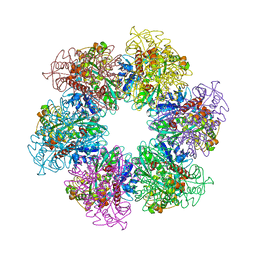 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: form two of GS-1 | | Descriptor: | GLUTAMINE, Glutamine synthetase, MAGNESIUM ION | | Authors: | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | Deposit date: | 2013-07-11 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
4LNI
 
 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: structure of the transition state complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutamine synthetase, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, ... | | Authors: | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | Deposit date: | 2013-07-11 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5793 Å) | | Cite: | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
1HAB
 
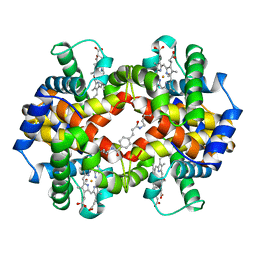 | | CROSSLINKED HAEMOGLOBIN | | Descriptor: | 4-CARBOXYCINNAMIC ACID, CARBON MONOXIDE, HEMOGLOBIN A, ... | | Authors: | Schumacher, M.A, Dixon, M.M, Kluger, R, Jones, R.T, Brennan, R.G. | | Deposit date: | 1996-03-13 | | Release date: | 1997-11-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Allosteric intermediates indicate R2 is the liganded hemoglobin end state.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
4LSD
 
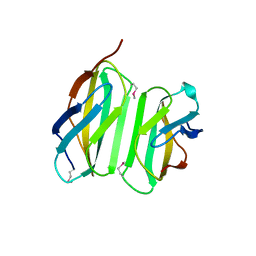 | | Myokine structure | | Descriptor: | Fibronectin type III domain-containing protein 5 | | Authors: | Schumacher, M.A, Ohashi, T, Shah, R.S, Chinnam, N, Erickson, H. | | Deposit date: | 2013-07-22 | | Release date: | 2013-10-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | The structure of irisin reveals a novel intersubunit beta-sheet fibronectin type III (FNIII) dimer: implications for receptor activation.
J.Biol.Chem., 288, 2013
|
|
1HAC
 
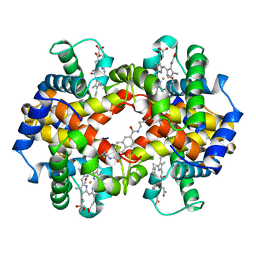 | | CROSSLINKED HAEMOGLOBIN | | Descriptor: | 2,6-DICARBOXYNAPHTHALENE, CARBON MONOXIDE, HEMOGLOBIN A, ... | | Authors: | Schumacher, M.A, Dixon, M.M, Kluger, R, Jones, R.T, Brennan, R.G. | | Deposit date: | 1996-03-13 | | Release date: | 1997-11-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Allosteric intermediates indicate R2 is the liganded hemoglobin end state.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
5U1G
 
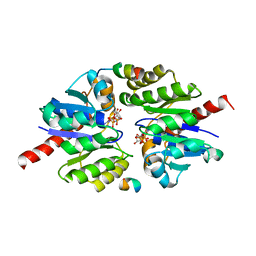 | | Structure of TP228 ParA-AMPPNP-ParB complex | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ParA, TP228 ParB fragment | | Authors: | Schumacher, M.A. | | Deposit date: | 2016-11-28 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Structures of partition protein ParA with nonspecific DNA and ParB effector reveal molecular insights into principles governing Walker-box DNA segregation.
Genes Dev., 31, 2017
|
|
5TZG
 
 | | Structure of the BldD CTD(D116A)-(c-di-GMP)2, form 2 | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DNA-binding protein, ZINC ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2016-11-21 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Streptomyces master regulator BldD binds c-di-GMP sequentially to create a functional BldD2-(c-di-GMP)4 complex.
Nucleic Acids Res., 45, 2017
|
|
5U1J
 
 | | Structure of pNOB8 ParA bound to nonspecific DNA | | Descriptor: | DNA (5'-D(*CP*GP*TP*GP*TP*AP*AP*TP*GP*AP*CP*GP*CP*CP*GP*GP*CP*GP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*CP*GP*CP*CP*GP*GP*CP*GP*TP*CP*AP*TP*GP*AP*CP*AP*CP*G)-3'), PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Schumacher, M.A. | | Deposit date: | 2016-11-28 | | Release date: | 2017-04-19 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structures of partition protein ParA with nonspecific DNA and ParB effector reveal molecular insights into principles governing Walker-box DNA segregation.
Genes Dev., 31, 2017
|
|
