1ZVV
 
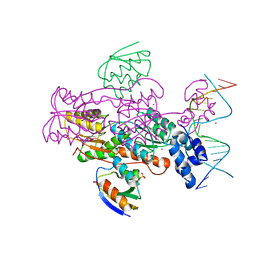 | | Crystal structure of a ccpa-crh-dna complex | | 分子名称: | DNA recognition strand CRE, Glucose-resistance amylase regulator, HPr-like protein crh, ... | | 著者 | Schumacher, M.A, Brennan, R.G, Hillen, W, Seidel, G. | | 登録日 | 2005-06-02 | | 公開日 | 2006-02-07 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Phosphoprotein Crh-Ser46-P displays altered binding to CcpA to effect carbon catabolite regulation.
J.Biol.Chem., 281, 2006
|
|
4GCK
 
 | | structure of no-dna complex | | 分子名称: | DNA (5'-D(*GP*TP*GP*AP*GP*TP*AP*CP*TP*CP*AP*C)-3'), Nucleoid occlusion factor SlmA | | 著者 | Schumacher, M.A. | | 登録日 | 2012-07-30 | | 公開日 | 2013-06-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4GFL
 
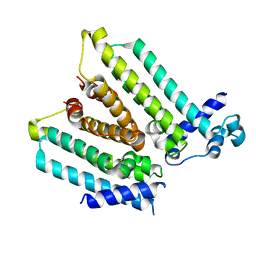 | | NO mechanism, slma | | 分子名称: | Nucleoid occlusion factor SlmA | | 著者 | Schumacher, M.A. | | 登録日 | 2012-08-03 | | 公開日 | 2013-06-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6UNX
 
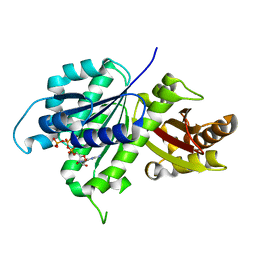 | | Structure of E. coli FtsZ(L178E)-GTP complex | | 分子名称: | Cell division protein FtsZ, GUANOSINE-5'-TRIPHOSPHATE | | 著者 | Schumacher, M.A. | | 登録日 | 2019-10-13 | | 公開日 | 2020-02-05 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | High-resolution crystal structures of Escherichia coli FtsZ bound to GDP and GTP.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4LNN
 
 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: structure of apo form of GS | | 分子名称: | Glutamine synthetase, MAGNESIUM ION, SULFATE ION | | 著者 | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | 登録日 | 2013-07-11 | | 公開日 | 2013-11-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
4LNI
 
 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: structure of the transition state complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Glutamine synthetase, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, ... | | 著者 | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | 登録日 | 2013-07-11 | | 公開日 | 2013-11-06 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.5793 Å) | | 主引用文献 | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
4LNO
 
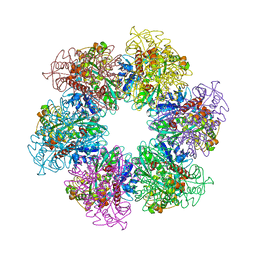 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: form two of GS-1 | | 分子名称: | GLUTAMINE, Glutamine synthetase, MAGNESIUM ION | | 著者 | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | 登録日 | 2013-07-11 | | 公開日 | 2013-11-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
4LNK
 
 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: structure of GS-glutamate-AMPPCP complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, GLUTAMIC ACID, Glutamine synthetase, ... | | 著者 | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | 登録日 | 2013-07-11 | | 公開日 | 2013-10-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.87 Å) | | 主引用文献 | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
3MKY
 
 | | Structure of SopB(155-323)-18mer DNA complex, I23 form | | 分子名称: | DNA (5'-D(*CP*TP*GP*GP*GP*AP*CP*CP*AP*TP*GP*GP*TP*CP*CP*CP*AP*G)-3'), Protein sopB, SULFATE ION | | 著者 | Schumacher, M.A, Piro, K, Xu, W. | | 登録日 | 2010-04-15 | | 公開日 | 2010-05-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.86 Å) | | 主引用文献 | Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes.
Nucleic Acids Res., 38, 2010
|
|
3MKW
 
 | | Structure of sopB(155-272)-18mer complex, I23 form | | 分子名称: | DNA (5'-D(*CP*TP*GP*GP*GP*AP*CP*CP*AP*TP*GP*GP*TP*CP*CP*CP*AP*G)-3'), Protein sopB, SULFATE ION | | 著者 | Schumacher, M.A, Piro, K, Xu, W. | | 登録日 | 2010-04-15 | | 公開日 | 2010-05-05 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes.
Nucleic Acids Res., 38, 2010
|
|
4LSD
 
 | | Myokine structure | | 分子名称: | Fibronectin type III domain-containing protein 5 | | 著者 | Schumacher, M.A, Ohashi, T, Shah, R.S, Chinnam, N, Erickson, H. | | 登録日 | 2013-07-22 | | 公開日 | 2013-10-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | The structure of irisin reveals a novel intersubunit beta-sheet fibronectin type III (FNIII) dimer: implications for receptor activation.
J.Biol.Chem., 288, 2013
|
|
6ALX
 
 | |
6UMK
 
 | | Structure of E. coli FtsZ(L178E)-GDP complex | | 分子名称: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | 著者 | Schumacher, M.A. | | 登録日 | 2019-10-09 | | 公開日 | 2020-02-05 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | High-resolution crystal structures of Escherichia coli FtsZ bound to GDP and GTP.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4GCT
 
 | | structure of No factor protein-DNA complex | | 分子名称: | DNA (5'-D(*TP*TP*AP*CP*GP*TP*GP*AP*GP*TP*AP*CP*TP*CP*AP*CP*GP*TP*AP*A)-3'), Nucleoid occlusion factor SlmA | | 著者 | Schumacher, M.A. | | 登録日 | 2012-07-30 | | 公開日 | 2013-06-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4OAY
 
 | | BldD CTD-c-di-GMP complex | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DNA-binding protein | | 著者 | Schumacher, M.A, Tschowri, N, Buttner, M, Brennan, R.G. | | 登録日 | 2014-01-06 | | 公開日 | 2014-11-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development.
Cell(Cambridge,Mass.), 158, 2014
|
|
1ZX4
 
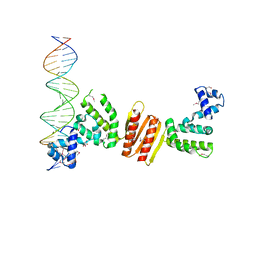 | | Structure of ParB bound to DNA | | 分子名称: | CITRIC ACID, Plasmid Partition par B protein, parS-small DNA centromere site | | 著者 | Schumacher, M.A, Funnell, B.E. | | 登録日 | 2005-06-06 | | 公開日 | 2005-11-29 | | 最終更新日 | 2017-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Structures of ParB bound to DNA reveal mechanism of partition complex formation.
Nature, 438, 2005
|
|
3M9A
 
 | |
1JUP
 
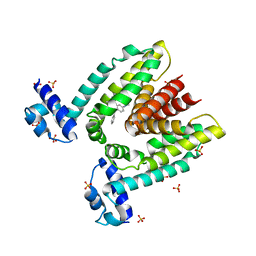 | | Crystal structure of the multidrug binding transcriptional repressor QacR bound to malachite green | | 分子名称: | HYPOTHETICAL TRANSCRIPTIONAL REGULATOR IN QACA 5'REGION, MALACHITE GREEN, SULFATE ION | | 著者 | Schumacher, M.A, Miller, M.C, Grkovic, S, Brown, M.H, Skurray, R.A, Brennan, R.G. | | 登録日 | 2001-08-24 | | 公開日 | 2001-12-12 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Structural mechanisms of QacR induction and multidrug recognition.
Science, 294, 2001
|
|
4GFK
 
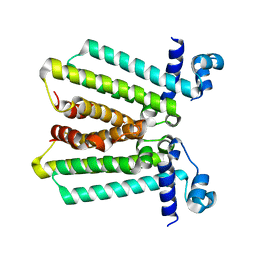 | | structures of NO factors | | 分子名称: | Nucleoid occlusion factor SlmA | | 著者 | Schumacher, M.A. | | 登録日 | 2012-08-03 | | 公開日 | 2013-06-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1JT0
 
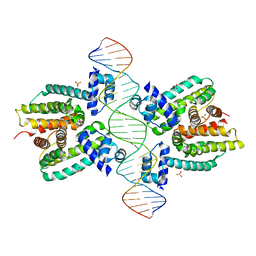 | | Crystal structure of a cooperative QacR-DNA complex | | 分子名称: | HYPOTHETICAL TRANSCRIPTIONAL REGULATOR IN QACA 5'REGION, QACA operator, SULFATE ION | | 著者 | Schumacher, M.A, Miller, M.C, Grkovic, S, Brown, M.H, Skurray, R.A, Brennan, R.G. | | 登録日 | 2001-08-20 | | 公開日 | 2002-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis for cooperative DNA binding by two dimers of the multidrug-binding protein QacR.
EMBO J., 21, 2002
|
|
1JTX
 
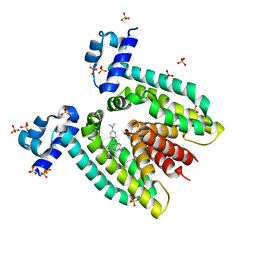 | | Crystal structure of the multidrug binding transcriptional regulator QacR bound to crystal violet | | 分子名称: | CRYSTAL VIOLET, HYPOTHETICAL TRANSCRIPTIONAL REGULATOR IN QACA 5'REGION, SULFATE ION | | 著者 | Schumacher, M.A, Miller, M.C, Grkovic, S, Brown, M.H, Skurray, R.A, Brennan, R.G. | | 登録日 | 2001-08-22 | | 公開日 | 2001-12-07 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structural mechanisms of QacR induction and multidrug recognition.
Science, 294, 2001
|
|
1JT6
 
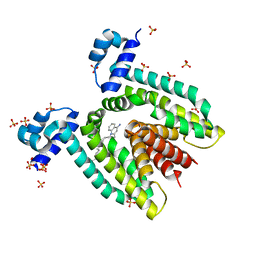 | | Crystal structure of the multidrug binding protein QacR bound to dequalinium | | 分子名称: | DEQUALINIUM, Hypothetical transcriptional regulator IN QACA 5'region, SULFATE ION | | 著者 | Schumacher, M.A, Miller, M.C, Grkovic, S, Brown, M.H, Skurray, R.A, Brennan, R.G. | | 登録日 | 2001-08-20 | | 公開日 | 2001-12-12 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | Structural mechanisms of QacR induction and multidrug recognition.
Science, 294, 2001
|
|
3MKZ
 
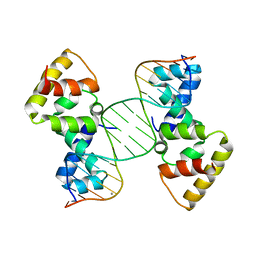 | | Structure of SopB(155-272)-18mer complex, P21 form | | 分子名称: | CALCIUM ION, DNA (5'-D(*CP*TP*GP*GP*GP*AP*CP*CP*AP*TP*GP*GP*TP*CP*CP*CP*AP*G)-3'), Protein sopB | | 著者 | Schumacher, M.A. | | 登録日 | 2010-04-15 | | 公開日 | 2010-05-05 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes.
Nucleic Acids Res., 38, 2010
|
|
3M8K
 
 | |
4FE4
 
 | | Crystal structure of apo E. coli XylR | | 分子名称: | Xylose operon regulatory protein | | 著者 | Schumacher, M.A, Ni, L. | | 登録日 | 2012-05-29 | | 公開日 | 2012-12-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.45 Å) | | 主引用文献 | Structures of the Escherichia coli transcription activator and regulator of diauxie, XylR: an AraC DNA-binding family member with a LacI/GalR ligand-binding domain.
Nucleic Acids Res., 41, 2013
|
|
