2UZ1
 
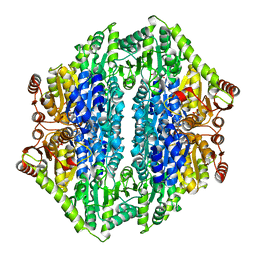 | | 1.65 Angstrom structure of Benzaldehyde Lyase complexed with 2-methyl- 2,4-pentanediol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BENZALDEHYDE LYASE, THIAMINE DIPHOSPHATE | | Authors: | Maraite, A, Schmidt, T, Ansorge-Schumacher, M.B, Brzozowski, A.M, Grogan, G. | | Deposit date: | 2007-04-23 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the Thdp-Dependent Enzyme Benzaldehyde Lyase Refined to 1.65 A Resolution.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
7QUY
 
 | | Alcohol Dehydrogenase from Thauera aromatica complexed with NADH | | Descriptor: | 6-hydroxycyclohex-1-ene-1-carbonyl-CoA dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Petchey, M.L, Stark, F, Ansorge-Schumacher, M, Grogan, G. | | Deposit date: | 2022-01-19 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Advanced Insights into Catalytic and Structural Features of the Zinc-Dependent Alcohol Dehydrogenase from Thauera aromatica.
Chembiochem, 23, 2022
|
|
7QUL
 
 | | Alcohol Dehydrogenase from Thauera aromatica K319A/K320A mutant | | Descriptor: | 1,2-ETHANEDIOL, 6-hydroxycyclohex-1-ene-1-carbonyl-CoA dehydrogenase, ZINC ION | | Authors: | Petchey, M.L, Stark, F, Ansorge-Schumacher, M, Grogan, G. | | Deposit date: | 2022-01-18 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Advanced Insights into Catalytic and Structural Features of the Zinc-Dependent Alcohol Dehydrogenase from Thauera aromatica.
Chembiochem, 23, 2022
|
|
6QHE
 
 | | Alcohol Dehydrogenase from Arthrobacter sp. TS-15 in complex with NAD+ | | Descriptor: | Alcohol Dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Lockie, C, Beloti, L, Shanati, T, Ansorge-Schumacher, M, Grogan, G. | | Deposit date: | 2019-01-16 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Two Enantiocomplementary Ephedrine Dehydrogenases from Arthrobacter sp. TS-15 with Broad Substrate Specificity
Acs Catalysis, 9, 2019
|
|
6WMR
 
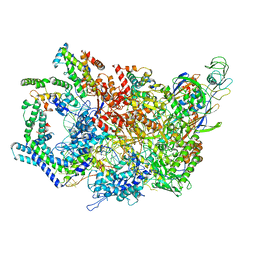 | | F. tularensis RNAPs70-(MglA-SspA)-iglA DNA complex | | Descriptor: | DNA NT-strand, DNA NT-strand downstream, DNA T-strand, ... | | Authors: | Travis, B.A, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2020-04-21 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural Basis for Virulence Activation of Francisella tularensis.
Mol.Cell, 81, 2021
|
|
6WMU
 
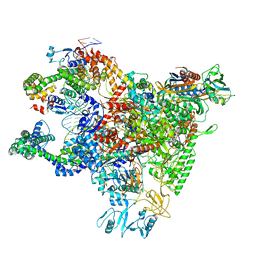 | | E. coli RNAPs70-SspA-gadA DNA complex | | Descriptor: | DNA NT-strand, DNA NT-strand downstream, DNA T-strand, ... | | Authors: | Travis, B.A, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2020-04-21 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural Basis for Virulence Activation of Francisella tularensis.
Mol.Cell, 81, 2021
|
|
6WMP
 
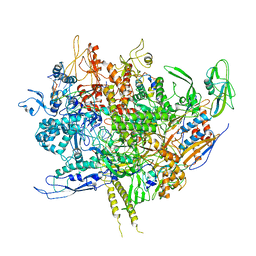 | | F. tularensis RNAPs70-iglA DNA complex | | Descriptor: | DNA NT-strand, DNA T-strand, DNA-directed RNA polymerase subunit alpha 1, ... | | Authors: | Travis, B.A, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2020-04-21 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural Basis for Virulence Activation of Francisella tularensis.
Mol.Cell, 81, 2021
|
|
5HW8
 
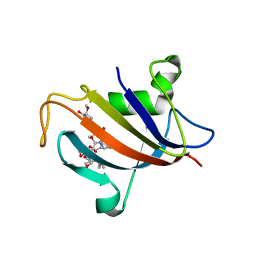 | |
5HW6
 
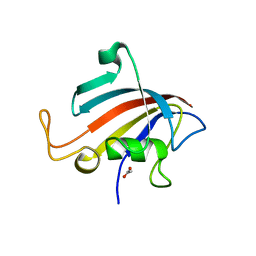 | |
4C4O
 
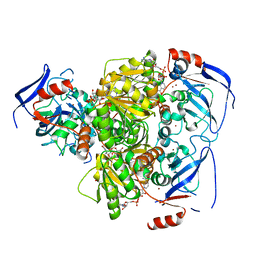 | | Structure of carbonyl reductase CPCR2 from Candida parapsilosis in complex with NADH | | Descriptor: | 1,2-ETHANEDIOL, CARBONYL REDUCTASE CPCR2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Man, H, Loderer, C, Ansorge-Schumacher, M, Grogan, G. | | Deposit date: | 2013-09-06 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of Nadh-Dependent Carbonyl Reductase (Cpcr2) from Candida Parapsilosis Provides Insight Into Mutations that Improve Catalytic Properties
Chemcatchem, 6, 2014
|
|
5HW7
 
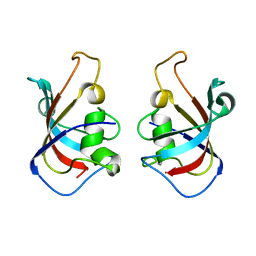 | |
5HWC
 
 | |
5HSZ
 
 | |
4YIX
 
 | | Structure of MRB1590 bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MERCURY (II) ION, ... | | Authors: | Shaw, P.L.R, Schumacher, M.A. | | Deposit date: | 2015-03-02 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the T. brucei kRNA editing factor MRB1590 reveal unique RNA-binding pore motif contained within an ABC-ATPase fold.
Nucleic Acids Res., 43, 2015
|
|
4D8J
 
 | | Structure of E. coli MatP-mats complex | | Descriptor: | 5'-D(*TP*TP*CP*GP*TP*GP*AP*CP*AP*AP*TP*GP*TP*CP*AP*CP*GP*AP*A)-3', 5'-D(*TP*TP*CP*GP*TP*GP*AP*CP*AP*TP*TP*GP*TP*CP*AP*CP*GP*AP*A)-3', Macrodomain Ter protein | | Authors: | Dupaigne, P, Tonthat, N.K, Espeli, O, Whitfill, T, Boccard, F, Schumacher, M.A. | | Deposit date: | 2012-01-10 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Molecular basis for a protein-mediated DNA-bridging mechanism that functions in condensation of the E. coli chromosome.
Mol.Cell, 48, 2012
|
|
6WMT
 
 | | F. tularensis RNAPs70-(MglA-SspA)-ppGpp-PigR-iglA DNA complex | | Descriptor: | DNA NT-strand, DNA NT-strand downstream, DNA T-strand, ... | | Authors: | Travis, B.A, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2020-04-21 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Structural Basis for Virulence Activation of Francisella tularensis.
Mol.Cell, 81, 2021
|
|
7TFB
 
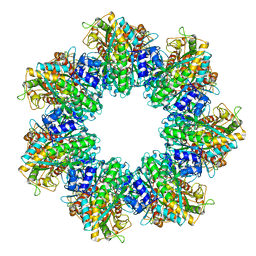 | | P. polymyxa GS(14)-Q-GlnR peptide | | Descriptor: | GLUTAMINE, GlnR C-tail peptide, Glutamine synthetase, ... | | Authors: | Travis, B.A, Peck, J, Schumacher, M.A. | | Deposit date: | 2022-01-06 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.28 Å) | | Cite: | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
7TF9
 
 | | L. monocytogenes GS(14)-Q-GlnR peptide | | Descriptor: | C-tail peptide of Glutamine synthetase repressor, GLUTAMINE, Glutamine synthetase, ... | | Authors: | Travis, B.A, Peck, J, Schumacher, M.A. | | Deposit date: | 2022-01-06 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
7TFA
 
 | | P. polymyxa GS(12)-Q-GlnR peptide | | Descriptor: | GLUTAMINE, GlnR C-tail peptide, Glutamine synthetase, ... | | Authors: | Travis, B.A, Peck, J, Schumacher, M.A. | | Deposit date: | 2022-01-06 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.07 Å) | | Cite: | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
7TF6
 
 | | S. aureus GS(12)-Q-GlnR peptide | | Descriptor: | GLUTAMINE, Glutamine synthetase, MAGNESIUM ION, ... | | Authors: | Travis, B.A, Peck, J, Schumacher, M.A. | | Deposit date: | 2022-01-06 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
7TFC
 
 | | B. subtilis GS(14)-Q-GlnR peptide | | Descriptor: | GLUTAMINE, GlnR C-tail peptide, Glutamine synthetase, ... | | Authors: | Travis, B.A, Peck, J, Schumacher, M.A. | | Deposit date: | 2022-01-06 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (1.96 Å) | | Cite: | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
4YIY
 
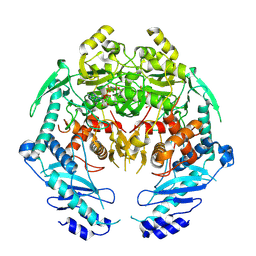 | | Structure of MRB1590 bound to AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, kRNA Editing A6 Specific Protein | | Authors: | Shaw, P.L.R, Schumacher, M.A. | | Deposit date: | 2015-03-02 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.016 Å) | | Cite: | Structures of the T. brucei kRNA editing factor MRB1590 reveal unique RNA-binding pore motif contained within an ABC-ATPase fold.
Nucleic Acids Res., 43, 2015
|
|
3ZQL
 
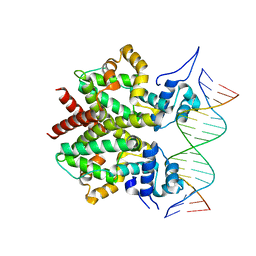 | | DNA-bound form of TetR-like repressor SimR | | Descriptor: | 5'-D(*DTP*TP*CP*GP*TP*AP*CP*GP*CP*CP*GP*TP*AP*DCP *GP*AP*A)-3', 5'-D(*DTP*TP*CP*GP*TP*AP*CP*GP*GP*CP*GP*TP*AP*DCP *GP*AP*A)-3', PUTATIVE REPRESSOR SIMREG2 | | Authors: | Le, T.B.K, Schumacher, M.A, Lawson, D.M, Brennan, R.G, Buttner, M.J. | | Deposit date: | 2011-06-10 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The Crystal Structure of the Tetr Family Transcriptional Repressor Simr Bound to DNA and the Role of a Flexible N-Terminal Extension in Minor Groove Binding.
Nucleic Acids Res., 39, 2011
|
|
5U56
 
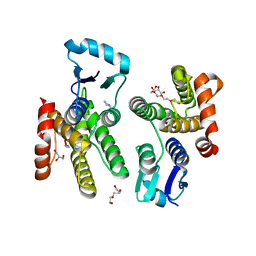 | | Structure of Francisella tularensis heterodimeric SspA (MglA-SspA) | | Descriptor: | GLYCEROL, Macrophage growth locus A, PENTAETHYLENE GLYCOL, ... | | Authors: | Cuthbert, B.J, Schumacher, M.A, Brennan, R.G. | | Deposit date: | 2016-12-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Dissection of the molecular circuitry controlling virulence in Francisella tularensis.
Genes Dev., 31, 2017
|
|
5U51
 
 | | Structure of Francisella tularensis heterodimeric SspA (MglA-SspA) in complex with ppGpp | | Descriptor: | GLYCEROL, GUANOSINE-5',3'-TETRAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Cuthbert, B.J, Schumacher, M.A, Brennan, R.G. | | Deposit date: | 2016-12-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dissection of the molecular circuitry controlling virulence in Francisella tularensis.
Genes Dev., 31, 2017
|
|
