8TGE
 
 | |
8UFJ
 
 | | Structure of M. mazei GS(R167L-A168G) apo form | | Descriptor: | Glutamine synthetase, MAGNESIUM ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2023-10-04 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | M. mazei glutamine synthetase and glutamine synthetase-GlnK1 structures reveal enzyme regulation by oligomer modulation.
Nat Commun, 14, 2023
|
|
7TZV
 
 | | Structure of DriD C-domain bound to 9mer ssDNA | | Descriptor: | DNA (5'-D(*TP*AP*GP*TP*CP*TP*AP*CP*T)-3'), WYL domain-containing protein | | Authors: | Schumacher, M.A, Laub, M. | | Deposit date: | 2022-02-16 | | Release date: | 2022-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | ssDNA is an allosteric regulator of the C. crescentus SOS-independent DNA damage response transcription activator, DriD.
Genes Dev., 36, 2022
|
|
8BAR
 
 | |
8BAS
 
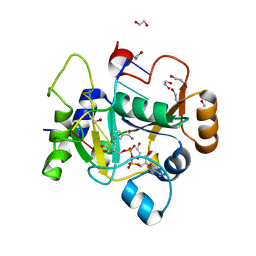 | | E. coli C7 DarT1 in complex with carba-NAD and DNA | | Descriptor: | 1,2-ETHANEDIOL, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, DNA (5'-D(*AP*AP*GP*AP*C)-3'), ... | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2022-10-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Molecular basis for the reversible ADP-ribosylation of guanosine bases.
Mol.Cell, 83, 2023
|
|
6U9X
 
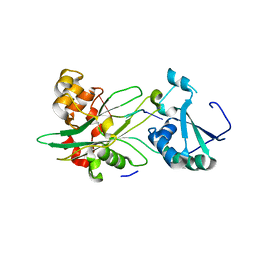 | | Structure of T. brucei MERS1-RNA complex | | Descriptor: | Mitochondrial edited mRNA stability factor 1, RNA (5'-R(*GP*AP*GP*AP*GP*GP*GP*GP*GP*UP*U)-3') | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-09-09 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of MERS1, the 5' processing enzyme of mitochondrial mRNAs inTrypanosoma brucei.
Rna, 26, 2020
|
|
6AMA
 
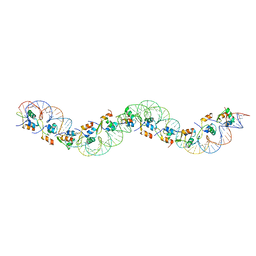 | |
8SVA
 
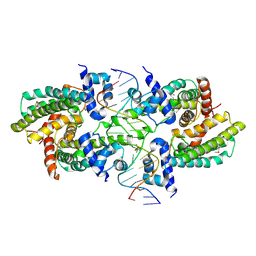 | | Structure of the Rhodococcus sp. USK13 DarR-20 bp DNA complex | | Descriptor: | DNA (5'-D(*TP*AP*GP*AP*TP*AP*CP*TP*CP*CP*GP*GP*AP*GP*TP*AP*TP*CP*TP*A)-3'), PHOSPHATE ION, TetR/AcrR family transcriptional regulator | | Authors: | Schumacher, M.A. | | Deposit date: | 2023-05-15 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structures of the DarR transcription regulator reveal unique modes of second messenger and DNA binding.
Nat Commun, 14, 2023
|
|
8SUA
 
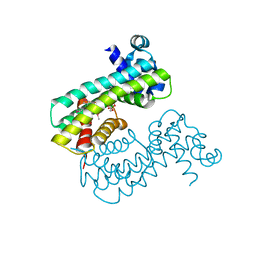 | | Structure of M. baixiangningiae DarR-ligand complex | | Descriptor: | 3-azanyl-3-(hydroxymethyl)-1,5,7,11-tetraoxa-6$l^{4}-boraspiro[5.5]undecan-9-ol, DarR | | Authors: | Schumacher, M.A. | | Deposit date: | 2023-05-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of the DarR transcription regulator reveal unique modes of second messenger and DNA binding.
Nat Commun, 14, 2023
|
|
5IXL
 
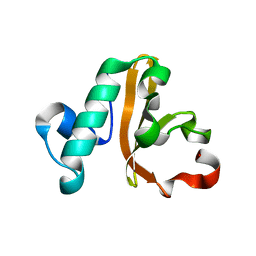 | | Structure of P. vulgaris HigB toxin Y91A variant | | Descriptor: | CHLORIDE ION, Endoribonuclease HigB | | Authors: | Schureck, M.A, Repack, A.A, Miles, S.J, Marquez, J, Dunham, C.M. | | Deposit date: | 2016-03-23 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanism of endonuclease cleavage by the HigB toxin.
Nucleic Acids Res., 44, 2016
|
|
5K58
 
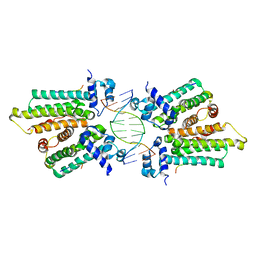 | |
7TEA
 
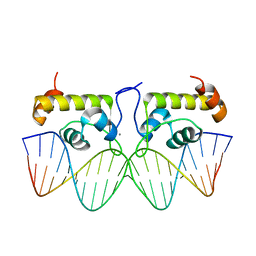 | | Crystal structure of S. aureus GlnR-DNA complex | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*GP*TP*GP*TP*CP*AP*GP*AP*TP*AP*AP*TP*CP*TP*GP*AP*CP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*GP*TP*CP*AP*GP*AP*TP*TP*AP*TP*CP*TP*GP*AP*CP*AP*CP*G)-3'), ... | | Authors: | Schumacher, M.A. | | Deposit date: | 2022-01-04 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
7TDP
 
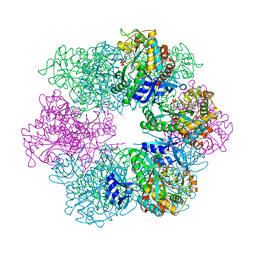 | |
7TDV
 
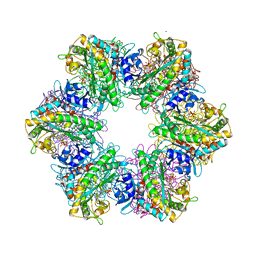 | |
7TEN
 
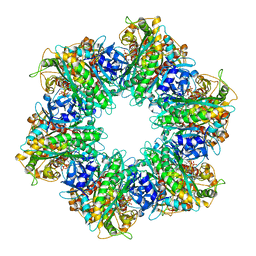 | |
5KBJ
 
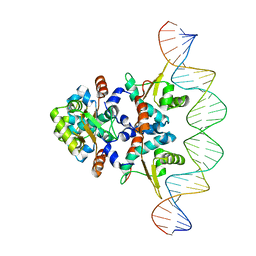 | | Structure of Rep-DNA complex | | Descriptor: | DNA (32-MER), Replication initiator A, N-terminal | | Authors: | Schumacher, M. | | Deposit date: | 2016-06-03 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Mechanism of staphylococcal multiresistance plasmid replication origin assembly by the RepA protein.
Proc. Natl. Acad. Sci. U.S.A., 111, 2014
|
|
8BAT
 
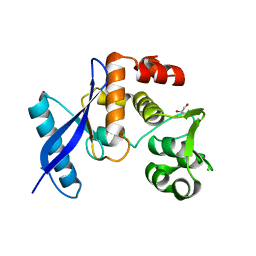 | | Geobacter lovleyi NADAR | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Geobacter lovleyi NADAR | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2022-10-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the reversible ADP-ribosylation of guanosine bases.
Mol.Cell, 83, 2023
|
|
8DPK
 
 | |
6AMK
 
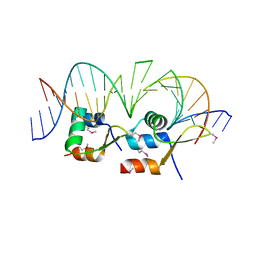 | | Structure of Streptomyces venezuelae BldC-whiI opt complex | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*TP*CP*CP*GP*AP*AP*TP*TP*AP*CP*CP*CP*GP*AP*AP*TP*TP*G)-3'), DNA (5'-D(*TP*TP*CP*AP*AP*TP*TP*CP*GP*GP*GP*TP*AP*AP*TP*TP*CP*GP*GP*GP*CP*A)-3'), Putative DNA-binding protein | | Authors: | Schumacher, M.A. | | Deposit date: | 2017-08-09 | | Release date: | 2018-03-28 | | Last modified: | 2018-11-07 | | Method: | X-RAY DIFFRACTION (3.288 Å) | | Cite: | The MerR-like protein BldC binds DNA direct repeats as cooperative multimers to regulate Streptomyces development.
Nat Commun, 9, 2018
|
|
8SUK
 
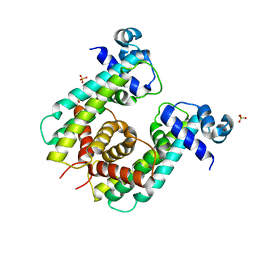 | | Structure of Rhodococcus sp. USK13 DarR-c-di-AMP complex | | Descriptor: | DNA (5'-D(*AP*A)-3'), DarR, SULFATE ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2023-05-12 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures of the DarR transcription regulator reveal unique modes of second messenger and DNA binding.
Nat Commun, 14, 2023
|
|
8SV6
 
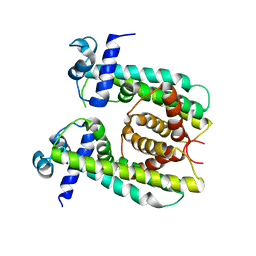 | | Structure of the M. smegmatis DarR protein | | Descriptor: | Fatty acid metabolism regulator protein | | Authors: | Schumacher, M.A. | | Deposit date: | 2023-05-15 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Structures of the DarR transcription regulator reveal unique modes of second messenger and DNA binding.
Nat Commun, 14, 2023
|
|
8CSH
 
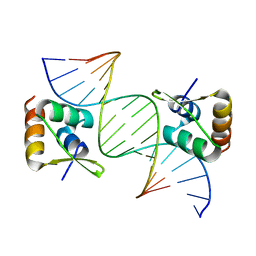 | |
5IWH
 
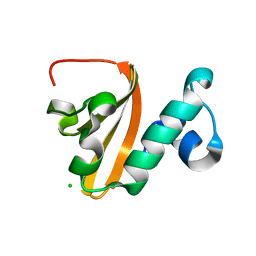 | | Structure of P. vulgaris HigB toxin delta H92 | | Descriptor: | CHLORIDE ION, Endoribonuclease HigB | | Authors: | Schureck, M.A, Repack, A.A, Miles, S.J, Marquez, J, Dunham, C.M. | | Deposit date: | 2016-03-22 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Mechanism of endonuclease cleavage by the HigB toxin.
Nucleic Acids Res., 44, 2016
|
|
7KIY
 
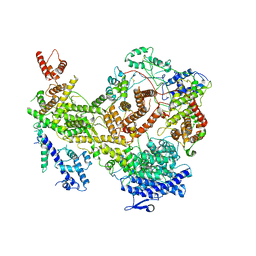 | | Plasmodium falciparum RhopH complex in soluble form | | Descriptor: | Cytoadherence linked asexual protein 3, High molecular weight rhoptry protein 3, High molecular weight rhoptry protein-2 | | Authors: | Schureck, M.A, Darling, J.E, Merk, A, Subramaniam, S, Desai, S.A. | | Deposit date: | 2020-10-25 | | Release date: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Malaria parasites use a soluble RhopH complex for erythrocyte invasion and an integral form for nutrient uptake.
Elife, 10, 2021
|
|
6P5R
 
 | | Structure of T. brucei MERS1-GDP complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Mitochondrial edited mRNA stability factor 1 | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-05-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures of MERS1, the 5' processing enzyme of mitochondrial mRNAs inTrypanosoma brucei.
Rna, 26, 2020
|
|
