3KZ5
 
 | | Structure of cdomain | | 分子名称: | ACETATE ION, Protein sopB | | 著者 | Schumacher, M.A. | | 登録日 | 2009-12-07 | | 公開日 | 2010-03-31 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes.
Nucleic Acids Res., 38, 2010
|
|
1QX7
 
 | |
3MKZ
 
 | | Structure of SopB(155-272)-18mer complex, P21 form | | 分子名称: | CALCIUM ION, DNA (5'-D(*CP*TP*GP*GP*GP*AP*CP*CP*AP*TP*GP*GP*TP*CP*CP*CP*AP*G)-3'), Protein sopB | | 著者 | Schumacher, M.A. | | 登録日 | 2010-04-15 | | 公開日 | 2010-05-05 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes.
Nucleic Acids Res., 38, 2010
|
|
5HTG
 
 | |
1ZX4
 
 | | Structure of ParB bound to DNA | | 分子名称: | CITRIC ACID, Plasmid Partition par B protein, parS-small DNA centromere site | | 著者 | Schumacher, M.A, Funnell, B.E. | | 登録日 | 2005-06-06 | | 公開日 | 2005-11-29 | | 最終更新日 | 2017-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Structures of ParB bound to DNA reveal mechanism of partition complex formation.
Nature, 438, 2005
|
|
1QX5
 
 | |
1KQ1
 
 | | 1.55 A Crystal structure of the pleiotropic translational regulator, Hfq | | 分子名称: | ACETIC ACID, Host Factor for Q beta | | 著者 | Schumacher, M.A, Pearson, R.F, Moller, T, Valentin-Hansen, P, Brennan, R.G. | | 登録日 | 2002-01-03 | | 公開日 | 2002-07-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein.
EMBO J., 21, 2002
|
|
4R25
 
 | | Structure of B. subtilis GlnK | | 分子名称: | Nitrogen regulatory PII-like protein, ZINC ION | | 著者 | Schumacher, M.A. | | 登録日 | 2014-08-08 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.5193 Å) | | 主引用文献 | Structures of regulatory machinery reveal novel molecular mechanisms controlling B. subtilis nitrogen homeostasis.
Genes Dev., 29, 2015
|
|
3PM1
 
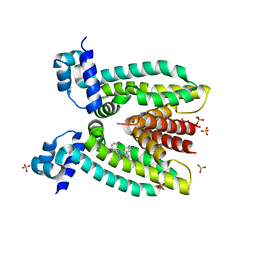 | | Structure of QacR E90Q bound to Ethidium | | 分子名称: | ETHIDIUM, HTH-type transcriptional regulator qacR, SULFATE ION | | 著者 | Schumacher, M.A. | | 登録日 | 2010-11-15 | | 公開日 | 2011-07-13 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A single acidic residue can guide binding site selection but does not govern QacR cationic-drug affinity.
Plos One, 6, 2011
|
|
1SXH
 
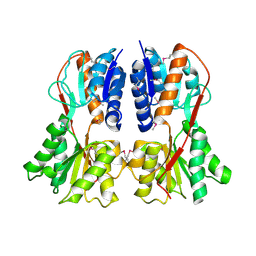 | | apo structure of B. megaterium transcription regulator | | 分子名称: | Glucose-resistance amylase regulator | | 著者 | Schumacher, M.A, Allen, G.S, Diel, M, Seidel, G, Hillen, W, Brennan, R.G. | | 登録日 | 2004-03-30 | | 公開日 | 2004-10-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural studies on the apo transcription factor form B. megaterium
Cell(Cambridge,Mass.), 118, 2004
|
|
4LNK
 
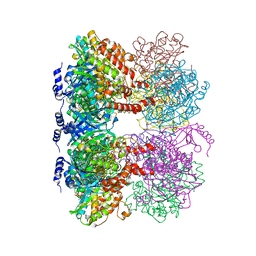 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: structure of GS-glutamate-AMPPCP complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, GLUTAMIC ACID, Glutamine synthetase, ... | | 著者 | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | 登録日 | 2013-07-11 | | 公開日 | 2013-10-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.87 Å) | | 主引用文献 | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
4LNN
 
 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: structure of apo form of GS | | 分子名称: | Glutamine synthetase, MAGNESIUM ION, SULFATE ION | | 著者 | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | 登録日 | 2013-07-11 | | 公開日 | 2013-11-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
4LNI
 
 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: structure of the transition state complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Glutamine synthetase, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, ... | | 著者 | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | 登録日 | 2013-07-11 | | 公開日 | 2013-11-06 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.5793 Å) | | 主引用文献 | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
4LNO
 
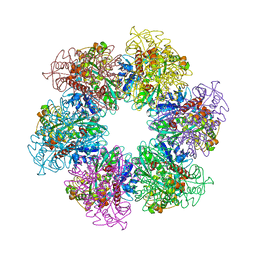 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: form two of GS-1 | | 分子名称: | GLUTAMINE, Glutamine synthetase, MAGNESIUM ION | | 著者 | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | 登録日 | 2013-07-11 | | 公開日 | 2013-11-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
8C1A
 
 | |
8C19
 
 | | SARS-CoV-2 NSP3 macrodomain in complex with 1-methyl-4-[5-(morpholin-4-ylcarbonyl)-2-furyl]-1H-pyrrolo[2,3-b]pyridine | | 分子名称: | 1,2-ETHANEDIOL, Non-structural protein 3, [5-(1-methylpyrrolo[2,3-b]pyridin-4-yl)furan-2-yl]-morpholin-4-yl-methanone | | 著者 | Schuller, M, Ahel, I. | | 登録日 | 2022-12-20 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Discovery and Development Strategies for SARS-CoV-2 NSP3 Macrodomain Inhibitors.
Pathogens, 12, 2023
|
|
5I41
 
 | | Structure of the apo RacA DNA binding domain | | 分子名称: | Chromosome-anchoring protein RacA | | 著者 | schumacher, M.A. | | 登録日 | 2016-02-11 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Molecular insights into DNA binding and anchoring by the Bacillus subtilis sporulation kinetochore-like RacA protein.
Nucleic Acids Res., 44, 2016
|
|
3DNV
 
 | | MDT Protein | | 分子名称: | DNA (5'-D(*DAP*DCP*DTP*DAP*DTP*DCP*DCP*DCP*DCP*DTP*DTP*DAP*DAP*DGP*DGP*DGP*DGP*DAP*DTP*DAP*DG)-3'), HTH-type transcriptional regulator hipB, Protein hipA, ... | | 著者 | schumacher, M.A. | | 登録日 | 2008-07-02 | | 公開日 | 2009-01-27 | | 最終更新日 | 2023-04-05 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB.
Science, 323, 2009
|
|
5KOA
 
 | |
1SLJ
 
 | | Solution structure of the S1 domain of RNase E from E. coli | | 分子名称: | Ribonuclease E | | 著者 | Schubert, M, Edge, R.E, Lario, P, Cook, M.A, Strynadka, N.C.J, Mackie, G.A, McIntosh, L.P. | | 登録日 | 2004-03-05 | | 公開日 | 2004-08-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces.
J.Mol.Biol., 341, 2004
|
|
1QVT
 
 | |
5K1Y
 
 | |
3M8K
 
 | |
4LSD
 
 | | Myokine structure | | 分子名称: | Fibronectin type III domain-containing protein 5 | | 著者 | Schumacher, M.A, Ohashi, T, Shah, R.S, Chinnam, N, Erickson, H. | | 登録日 | 2013-07-22 | | 公開日 | 2013-10-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | The structure of irisin reveals a novel intersubunit beta-sheet fibronectin type III (FNIII) dimer: implications for receptor activation.
J.Biol.Chem., 288, 2013
|
|
4R4E
 
 | | Structure of GlnR-DNA complex | | 分子名称: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, DNA (5'-D(*AP*TP*TP*CP*TP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*AP*GP*TP*A)-3'), ... | | 著者 | Schumacher, M.A. | | 登録日 | 2014-08-19 | | 公開日 | 2015-03-04 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | Structures of regulatory machinery reveal novel molecular mechanisms controlling B. subtilis nitrogen homeostasis.
Genes Dev., 29, 2015
|
|
