4B9U
 
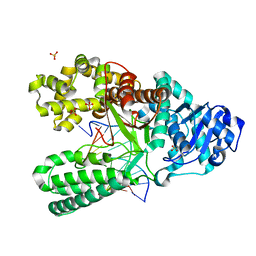 | | Structure of the high fidelity DNA polymerase I with an oxidative formamidopyrimidine-dG DNA lesion -dA basepair in the post-insertion site. | | Descriptor: | 5'-D(*CP*AP*AP*FOXP*CP*GP*AP*GP*TP*CP*AP*GP*GP*CP*TP)-3', 5'-D(*GP*CP*CP*TP*GP*AP*CP*TP*CP*GP*AP)-3', DNA POLYMERASE, ... | | Authors: | Gehrke, T.H, Lischke, U, Arnold, S, Schneider, S, Carell, T. | | Deposit date: | 2012-09-06 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected Non-Hoogsteen-Based Mutagenicity Mechanism of Fapy-DNA Lesions.
Nat.Chem.Biol., 9, 2013
|
|
4B9N
 
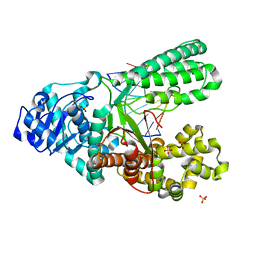 | | Structure of the high fidelity DNA polymerase I correctly bypassing the oxidative formamidopyrimidine-dA DNA lesion. | | Descriptor: | 5'-D(*CP*AP*AP*(FAX)*AP*GP*AP*GP*TP*CP*AP*GP*GP*CP*TP)-3', 5'-D(*GP*CP*CP*TP*GP*AP*CP*TP*CP*TP*TP*TP*TP)-3', DNA POLYMERASE, ... | | Authors: | Gehrke, T.H, Lischke, U, Arnold, S, Schneider, S, Carell, T. | | Deposit date: | 2012-09-05 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unexpected Non-Hoogsteen-Based Mutagenicity Mechanism of Fapy-DNA Lesions.
Nat.Chem.Biol., 9, 2013
|
|
4B9L
 
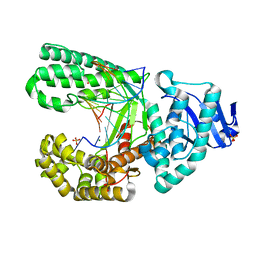 | | Structure of the high fidelity DNA polymerase I with the oxidative formamidopyrimidine-dA DNA lesion in the pre-insertion site. | | Descriptor: | 5'-D(*CP*AP*GP*FAX*AP*GP*AP*GP*TP*CP*AP*GP*GP*CP*TP)-3', 5'-D(*GP*CP*CP*TP*GP*AP*CP*TP*CP*TP)-3', DNA POLYMERASE I, ... | | Authors: | Gehrke, T.H, Lischke, U, Arnold, S, Schneider, S, Carell, T. | | Deposit date: | 2012-09-05 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Unexpected Non-Hoogsteen-Based Mutagenicity Mechanism of Fapy-DNA Lesions.
Nat.Chem.Biol., 9, 2013
|
|
4B9S
 
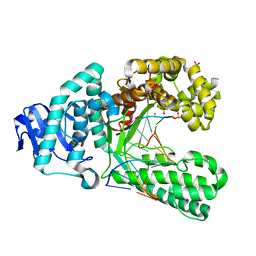 | | Structure of the high fidelity DNA polymerase I with an oxidative formamidopyrimidine-dG DNA lesion outside of the pre-insertion site. | | Descriptor: | 5'-D(*AP*CP*CP*TP*GP*AP*CP*TP*CP*TP)-3', 5'-D(*CP*AP*TP*FOXP*AP*GP*AP*GP*TP*CP*AP*GP*GP*TP*TP)-3', DNA POLYMERASE, ... | | Authors: | Gehrke, T.H, Lischke, U, Arnold, S, Schneider, S, Carell, T. | | Deposit date: | 2012-09-06 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Unexpected Non-Hoogsteen-Based Mutagenicity Mechanism of Fapy-DNA Lesions.
Nat.Chem.Biol., 9, 2013
|
|
4C5L
 
 | | Structure of the pyridoxal kinase from Staphylococcus aureus in complex with pyridoxal | | Descriptor: | 3-HYDROXY-5-(HYDROXYMETHYL)-2-METHYLISONICOTINALDEHYDE, 4,5-bis(hydroxymethyl)-2-methyl-pyridin-3-ol, PHOSPHOMETHYLPYRIMIDINE KINASE, ... | | Authors: | Nodwell, M, Alte, F, Sieber, S.A, Schneider, S. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Subfamily of Bacterial Ribokinases Utilizes a Hemithioacetal for Pyridoxal Phosphate Salvage.
J.Am.Chem.Soc., 136, 2014
|
|
4C5N
 
 | | Structure of the pyridoxal kinase from Staphylococcus aureus in complex with AMP-PCP and pyridoxal | | Descriptor: | 3-HYDROXY-5-(HYDROXYMETHYL)-2-METHYLISONICOTINALDEHYDE, 4,5-bis(hydroxymethyl)-2-methyl-pyridin-3-ol, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Nodwell, M, Alte, F, Sieber, S.A, Schneider, S. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Subfamily of Bacterial Ribokinases Utilizes a Hemithioacetal for Pyridoxal Phosphate Salvage.
J.Am.Chem.Soc., 136, 2014
|
|
4C5K
 
 | | Structure of the pyridoxal kinase from Staphylococcus aureus in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHOMETHYLPYRIMIDINE KINASE, SULFATE ION | | Authors: | Nodwell, M, Alte, F, Sieber, S.A, Schneider, S. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Subfamily of Bacterial Ribokinases Utilizes a Hemithioacetal for Pyridoxal Phosphate Salvage.
J.Am.Chem.Soc., 136, 2014
|
|
4C5J
 
 | | Structure of the pyridoxal kinase from Staphylococcus aureus | | Descriptor: | PHOSPHOMETHYLPYRIMIDINE KINASE, SULFATE ION | | Authors: | Nodwell, M, Alte, F, Sieber, S.A, Schneider, S. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Subfamily of Bacterial Ribokinases Utilizes a Hemithioacetal for Pyridoxal Phosphate Salvage.
J.Am.Chem.Soc., 136, 2014
|
|
4CH5
 
 | | Structure of pyrrolysyl-tRNA synthetase in complex with adenylated propionyl lysine | | Descriptor: | (S)-2-amino-6-propionamidohexanoic(((2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl phosphoric) anhydride, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Fluegel, V, Vrabel, M, Schneider, S. | | Deposit date: | 2013-11-28 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Site-Specific Incorporation of Lysine Derivatives Into Proteins.
Plos One, 9, 2014
|
|
4CH3
 
 | | Structure of pyrrolysyl-tRNA synthetase in complex with adenylated butyryl lysine | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-({[(2R)-2-amino-6-(butanoylamino)hexanoyl]oxy}phosphinato)adenosine, MAGNESIUM ION, ... | | Authors: | Fluegel, V, Vrabel, M, Schneider, S. | | Deposit date: | 2013-11-28 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Basis for the Site-Specific Incorporation of Lysine Derivatives Into Proteins.
Plos One, 9, 2014
|
|
4CH4
 
 | | Structure of pyrrolysyl-tRNA synthetase in complex with adenylated crotonyl lysine | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[({(2R)-2-amino-6-[(2E)-but-2-enoylamino]hexanoyl}oxy)phosphinato]adenosine, MAGNESIUM ION, ... | | Authors: | Fluegel, V, Vrabel, M, Schneider, S. | | Deposit date: | 2013-11-28 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural Basis for the Site-Specific Incorporation of Lysine Derivatives Into Proteins.
Plos One, 9, 2014
|
|
4CH6
 
 | | Structure of pyrrolysyl-tRNA synthetase in complex with adenylated propargyloxycarbonyl lysine | | Descriptor: | (S)-2-amino-6-(((prop-2-yn-1-yloxy)carbonyl)amino)hexanoic (((2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl phosphoric)anhydride, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Fluegel, V, Vrabel, M, Schneider, S. | | Deposit date: | 2013-11-28 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for the Site-Specific Incorporation of Lysine Derivatives Into Proteins.
Plos One, 9, 2014
|
|
4C5M
 
 | | Structure of the pyridoxal kinase from Staphylococcus aureus in complex with AMP-PCP | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, PHOSPHOMETHYLPYRIMIDINE KINASE, SULFATE ION | | Authors: | Nodwell, M, Alte, F, Sieber, S.A, Schneider, S. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Subfamily of Bacterial Ribokinases Utilizes a Hemithioacetal for Pyridoxal Phosphate Salvage.
J.Am.Chem.Soc., 136, 2014
|
|
6HLF
 
 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant - K32A | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, R-specific alcohol dehydrogenase | | Authors: | Hermann, J, Nowotny, P, Schneider, S, Hekmat, D, Weuster-Botz, D. | | Deposit date: | 2018-09-11 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Rational Crystal Contact Engineering of Lactobacillus brevis Alcohol Dehydrogenase To Promote Technical Protein Crystallization
Cryst.Growth Des., 2019
|
|
6H07
 
 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, R-specific alcohol dehydrogenase | | Authors: | Hermann, J, Nowotny, P, Biggel, P, Schneider, S, Hekmat, D, Weuster-Botz, D. | | Deposit date: | 2018-07-06 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Neutron and X-ray crystal structures of Lactobacillus brevis alcohol dehydrogenase reveal new insights into hydrogen-bonding pathways.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
