4HU2
 
 | | Crystal structure of LdtMt2, a L,D-transpeptidase from Mycobacterium tuberculosis: domain A and B | | 分子名称: | PROBABLE CONSERVED LIPOPROTEIN LPPS, SULFATE ION | | 著者 | Both, D, Steiner, E, Lindqvist, Y, Schnell, R, Schneider, G. | | 登録日 | 2012-11-02 | | 公開日 | 2012-12-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Structure of LdtMt2, an L,D-transpeptidase from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1BII
 
 | | THE CRYSTAL STRUCTURE OF H-2DD MHC CLASS I IN COMPLEX WITH THE HIV-1 DERIVED PEPTIDE P18-110 | | 分子名称: | BETA-2 MICROGLOBULIN, DECAMERIC PEPTIDE, MHC CLASS I H-2DD | | 著者 | Achour, A, Persson, K, Harris, R.A, Sundback, J, Sentman, C.L, Lindqvist, Y, Schneider, G, Karre, K. | | 登録日 | 1998-06-11 | | 公開日 | 1998-10-14 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The crystal structure of H-2Dd MHC class I complexed with the HIV-1-derived peptide P18-I10 at 2.4 A resolution: implications for T cell and NK cell recognition.
Immunity, 9, 1998
|
|
4HUC
 
 | | Crystal structure of LdtMt2, a L,D-transpeptidase from Mycobacterium tuberculosis: domain B and C | | 分子名称: | ACETATE ION, PROBABLE CONSERVED LIPOPROTEIN LPPS, SODIUM ION | | 著者 | Both, D, Steiner, E, Lindqvist, Y, Schnell, R, Schneider, G. | | 登録日 | 2012-11-02 | | 公開日 | 2012-12-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Structure of LdtMt2, an L,D-transpeptidase from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3FGP
 
 | |
3DKI
 
 | |
3CQ6
 
 | |
4IWS
 
 | | Putative Aromatic Acid Decarboxylase | | 分子名称: | PA0254, SULFATE ION | | 著者 | Jacewicz, A, Izumi, A, Brunner, K, Schneider, G. | | 登録日 | 2013-01-24 | | 公開日 | 2013-05-29 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Insights into the UbiD Protein Family from the Crystal Structure of PA0254 from Pseudomonas aeruginosa.
Plos One, 8, 2013
|
|
1ZJ8
 
 | | Structure of Mycobacterium tuberculosis NirA protein | | 分子名称: | CHLORIDE ION, IRON/SULFUR CLUSTER, Probable ferredoxin-dependent nitrite reductase NirA, ... | | 著者 | Schnell, R, Sandalova, T, Hellman, U, Lindqvist, Y, Schneider, G, Structural Proteomics in Europe (SPINE) | | 登録日 | 2005-04-28 | | 公開日 | 2005-05-31 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Siroheme- and [Fe4-S4]-dependent NirA from Mycobacterium tuberculosis Is a Sulfite Reductase with a Covalent Cys-Tyr Bond in the Active Site
J.Biol.Chem., 280, 2005
|
|
1ZJ9
 
 | | Structure of Mycobacterium tuberculosis NirA protein | | 分子名称: | CHLORIDE ION, IRON/SULFUR CLUSTER, Probable ferredoxin-dependent nitrite reductase NirA, ... | | 著者 | Schnell, R, Sandalova, T, Hellman, U, Lindqvist, Y, Schneider, G. | | 登録日 | 2005-04-28 | | 公開日 | 2005-05-31 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Siroheme- and [Fe4-S4]-dependent NirA from Mycobacterium tuberculosis Is a Sulfite Reductase with a Covalent Cys-Tyr Bond in the Active Site
J.Biol.Chem., 280, 2005
|
|
4JB1
 
 | | Crystal structure of P. aeruginosa MurB in complex with NADP+ | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Chen, M.W, Lohkamp, B, Schnell, R, Lescar, J, Schneider, G. | | 登録日 | 2013-02-19 | | 公開日 | 2013-07-17 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Substrate Channel Flexibility in Pseudomonas aeruginosa MurB Accommodates Two Distinct Substrates.
Plos One, 8, 2013
|
|
3PBC
 
 | |
4JXB
 
 | | RipD (Rv1566c) from Mycobacterium tuberculosis: a non-catalytic NlpC/p60 domain protein, adaptation to peptidoglycan-binding function | | 分子名称: | ACETATE ION, Invasion-associated protein | | 著者 | Both, D, Steiner, E.M, Schnell, R, Schneider, G. | | 登録日 | 2013-03-28 | | 公開日 | 2013-10-30 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | RipD (Rv1566c) from Mycobacterium tuberculosis: adaptation of an NlpC/p60 domain to a non-catalytic peptidoglycan-binding function.
Biochem.J., 457, 2014
|
|
3PBI
 
 | |
4JAY
 
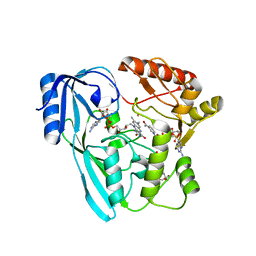 | | Crystal structure of P. aeruginosa MurB in complex with NADP+ | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Chen, M.W, Lohkamp, B, Schnell, R, Lescar, J, Schneider, G. | | 登録日 | 2013-02-19 | | 公開日 | 2013-07-17 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Substrate Channel Flexibility in Pseudomonas aeruginosa MurB Accommodates Two Distinct Substrates.
Plos One, 8, 2013
|
|
1XDS
 
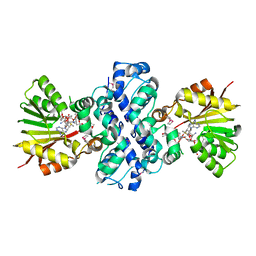 | | Crystal structure of Aclacinomycin-10-hydroxylase (RdmB) in complex with S-adenosyl-L-methionine (SAM) and 11-deoxy-beta-rhodomycin (DbrA) | | 分子名称: | 11-DEOXY-BETA-RHODOMYCIN, Protein RdmB, S-ADENOSYLMETHIONINE | | 著者 | Jansson, A, Koskiniemi, H, Erola, A, Wang, J, Mantsala, P, Schneider, G, Niemi, J, Structural Proteomics in Europe (SPINE) | | 登録日 | 2004-09-08 | | 公開日 | 2004-11-23 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Aclacinomycin 10-Hydroxylase Is a Novel Substrate-assisted Hydroxylase Requiring S-Adenosyl-L-methionine as Cofactor
J.Biol.Chem., 280, 2005
|
|
1XDU
 
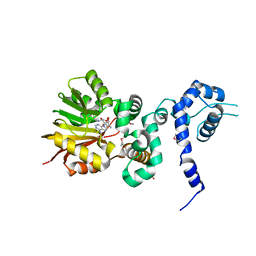 | | Crystal structure of Aclacinomycin-10-hydroxylase (RdmB) in complex with Sinefungin (SFG) | | 分子名称: | ACETATE ION, Protein RdmB, SINEFUNGIN | | 著者 | Jansson, A, Koskiniemi, H, Erola, A, Wang, J, Mantsala, P, Schneider, G, Niemi, J. | | 登録日 | 2004-09-08 | | 公開日 | 2004-11-23 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Aclacinomycin 10-Hydroxylase Is a Novel Substrate-assisted Hydroxylase Requiring S-Adenosyl-L-methionine as Cofactor
J.Biol.Chem., 280, 2005
|
|
3IHG
 
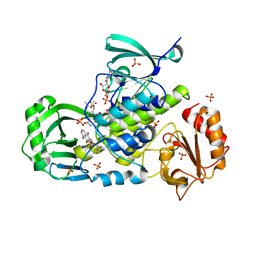 | | Crystal structure of a ternary complex of aklavinone-11 hydroxylase with FAD and aklavinone | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, RdmE, SULFATE ION, ... | | 著者 | Lindqvist, Y, Koskiniemi, H, Jansson, A, Sandalova, T, Schneider, G. | | 登録日 | 2009-07-30 | | 公開日 | 2009-09-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Structural basis for substrate recognition and specificity in aklavinone-11-hydroxylase from rhodomycin biosynthesis.
J.Mol.Biol., 393, 2009
|
|
3HT2
 
 | |
3HT1
 
 | |
1TRK
 
 | |
5AHK
 
 | | Crystal structure of acetohydroxy acid synthase Pf5 from Pseudomonas protegens | | 分子名称: | ACETOLACTATE SYNTHASE II, LARGE SUBUNIT, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Dobritzsch, D, Loschonsky, S, Mueller, M, Schneider, G. | | 登録日 | 2015-02-06 | | 公開日 | 2016-03-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | The Crystal Structure of the Acetohydroxy Acid Synthase Pf5 from Pseudomonas Protegens
To be Published
|
|
4RFV
 
 | | Structure of the Mycobacterium tuberculosis APS kinase CysC Cys556Ala mutant | | 分子名称: | Bifunctional enzyme CysN/CysC, PHOSPHATE ION | | 著者 | Poyraz, O, Brunner, K, Schnell, R, Schneider, G. | | 登録日 | 2014-09-28 | | 公開日 | 2015-01-28 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Crystal Structures of the Kinase Domain of the Sulfate-Activating Complex in Mycobacterium tuberculosis.
Plos One, 10, 2015
|
|
4AG3
 
 | | Crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with NADPH at 1.8A resolution | | 分子名称: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PENTAETHYLENE GLYCOL | | 著者 | Cukier, C.D, Schnell, R, Schneider, G, Lindqvist, Y. | | 登録日 | 2012-01-24 | | 公開日 | 2013-02-06 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4S1F
 
 | | Fructose-6-phosphate aldolase A from E.coli soaked in acetylacetone | | 分子名称: | Fructose-6-phosphate aldolase 1, pentane-2,4-dione | | 著者 | Stellmacher, L, Sandalova, T, Leptihn, S, Schneider, G, Sprenger, G.A, Samland, A.K. | | 登録日 | 2015-01-13 | | 公開日 | 2015-10-07 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.242 Å) | | 主引用文献 | Acid Base Catalyst Discriminates between a Fructose 6-Phosphate Aldolase and a Transaldolase
ChemCatChem, 2015
|
|
4RZ6
 
 | | Transaldolase B E96Q F178Y from E.coli | | 分子名称: | CHLORIDE ION, HEXAETHYLENE GLYCOL, SULFATE ION, ... | | 著者 | Stellmacher, L, Sandalova, T, Leptihn, S, Schneider, G, Sprenger, G.A, Samland, A.K. | | 登録日 | 2014-12-18 | | 公開日 | 2015-10-07 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.802 Å) | | 主引用文献 | Acid Base Catalyst Discriminates between a Fructose 6-Phosphate Aldolase and a Transaldolase
ChemCatChem, 2015
|
|
